Fig. 2 |. Three main classes of NAD+-consuming enzymes.
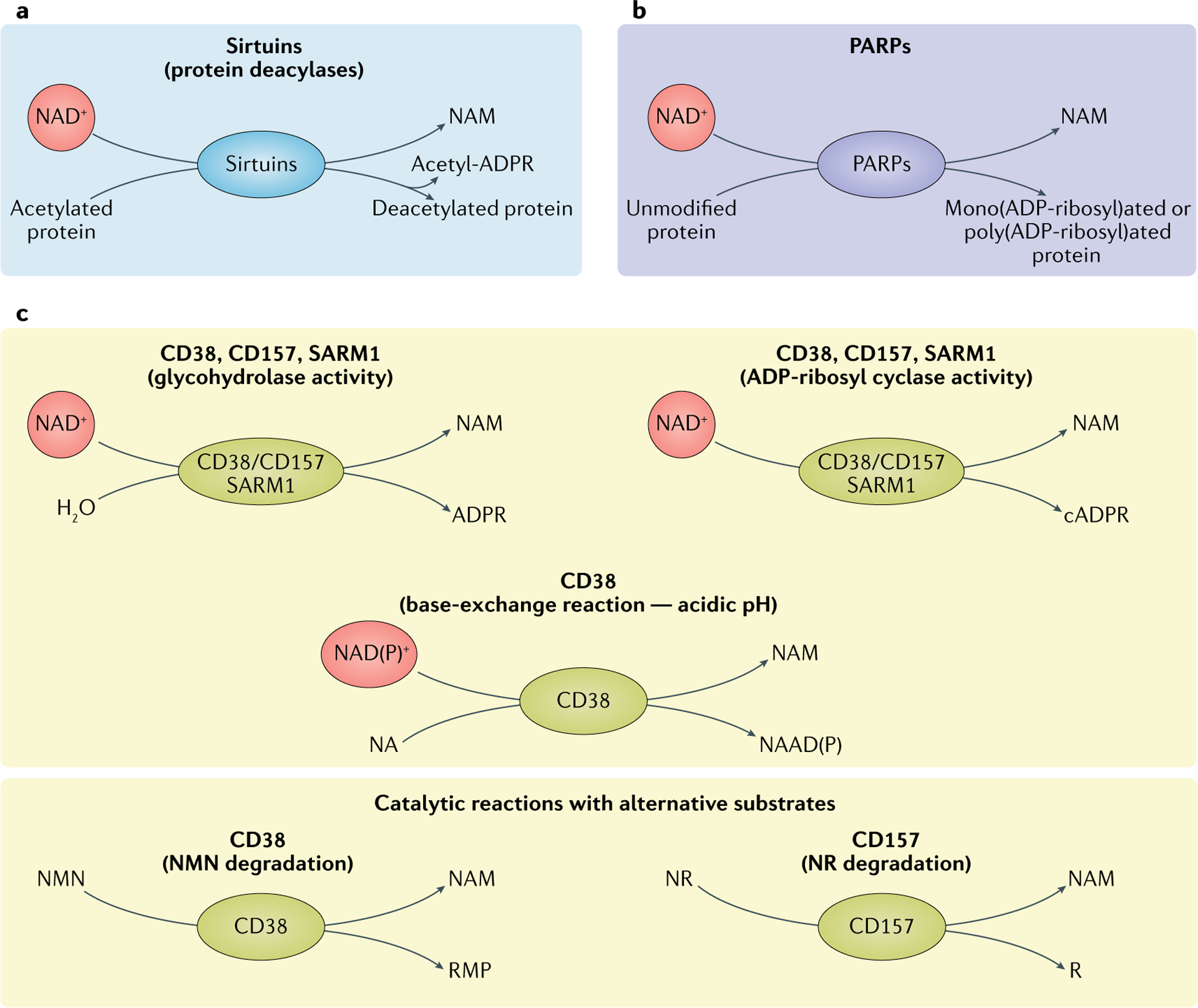
a | Sirtuins remove acyl groups from lysine residues on target proteins using nicotinamide adenine dinucleotide (NAD+) as a co-substrate. NAD+ is cleaved, generating nicotinamide (NAM) and ADP-ribose, where ADP-ribose serves as an acyl group acceptor, generating acetyl-ADP-ribose (acetyl-ADPR). b | Poly(ADP-ribose) polymerases (PARP1–PARP3) use NAD+ as a co-substrate to mono(ADP-ribosyl)ate or poly(ADP-ribosyl)ate target proteins, generating NAM as a by-product. c | Reactions of NAD+ glycohydrolases and cyclic ADP-ribose (cADPR) synthases (CD38, CD157 and SARM1). The main catalytic activity of this group of proteins is the hydrolysis of NAD+ to NAM and ADP-ribose. To a lesser extent, CD38, CD157 and SARM1 have ADP-ribosyl cyclase activity, generating NAM and cADPR from NAD+. In acidic conditions, CD38 can also perform a base-exchange reaction, swapping the NAM of NAD(P)+ for nicotinic acid (NA), generating nicotinic acid adenine dinucleotide (phosphate) (NAAD(P)). CD38 and CD157 are reported to be able to use alternative substrates in their catalytic reactions. CD38 can degrade NMN to NAM and ribose monophosphate (RMP), while CD157 can degrade NR, generating NAM and ribose (R). NR, nicotinamide riboside.
