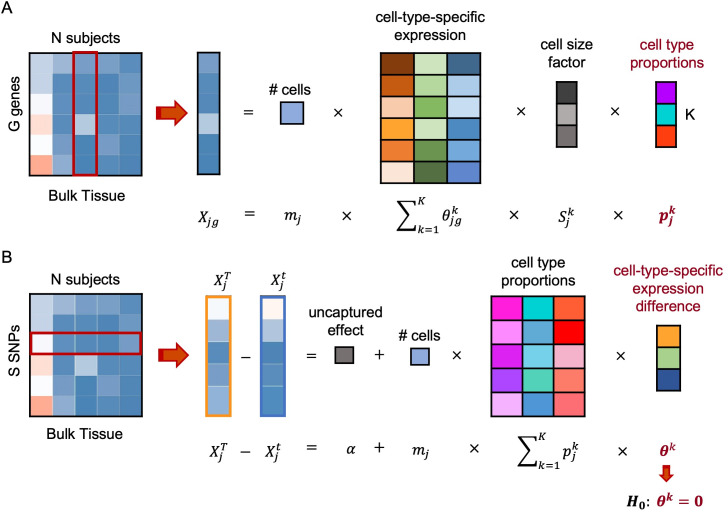
Fig 1. Overview of BSCET.
The method contains two steps. (A) First, we employ the deconvolution method, such as MuSiC [14], to infer the cell type proportion, , of the bulk tissue for each individual j of cell type k by incorporating the cell-type-specific expression information, , and cell size factor, , obtained from scRNA-seq data. (B) Second, at each transcribed SNP (tSNP) site, we align the tSNP alleles with respect to its reference and alternative alleles, T and t, across individuals, and model the expression difference between two allele-specific read counts, and , over the estimated cell type compositions, , through a linear model with an intercept term α to capture the information not explained by the selected K cell types, where both mj and are assumed known. The difference of population-level relative abundance between two transcribed alleles, θk, can therefore be inferred for each cell type. By testing for H0: θk = 0, we can detect cell-type-specific allelic expression imbalance across samples.