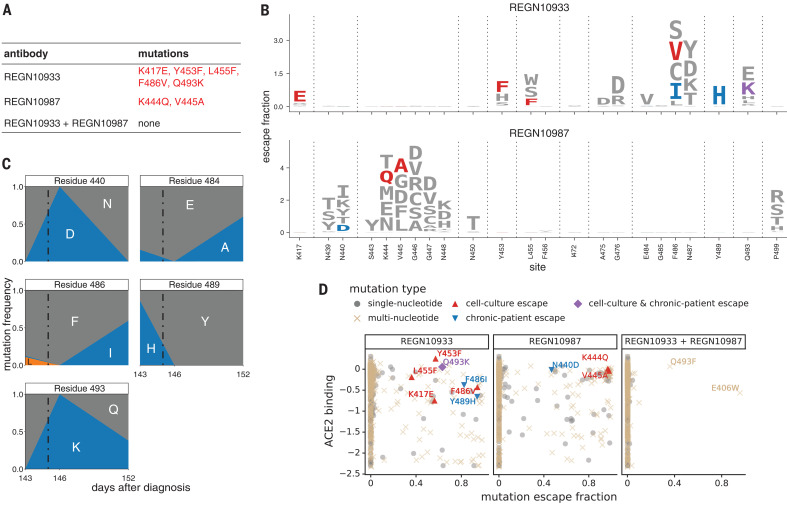
Fig. 2. Escape maps are consistent with viral mutations selected in cell culture and a persistently infected patient.
(A) Viral escape mutations selected by Regeneron with spike-pseudotyped VSV in cell culture in the presence of antibody (12). (B) Escape maps like those in Fig. 1A, but showing only mutations accessible by single-nucleotide changes to the Wuhan-Hu-1 sequence, with nongray colors indicating mutations in cell culture (red), the infected patient (blue), or both (purple). Figure S5 shows these maps colored by how mutations affect ACE2 affinity or RBD expression. (C) Dynamics of RBD mutations in a patient treated with REGN-COV2 at day 145 of infection (black dot-dash vertical line). E484A rose in frequency in linkage with F486I, but because E484A is not an escape mutation in our maps, it is not shown in other panels. See also fig. S4. (D) The escape mutations that arise in cell culture and the infected patient are single-nucleotide–accessible and escape antibody binding without imposing a large cost on ACE2 affinity [as measured using yeast display (7)]. Each point is a mutation, with shape and color indicating whether it is accessible and selected during viral growth. Points farther to the right on the x axis indicate stronger escape from antibody binding; points higher up on the y axis indicate higher ACE2 affinity.