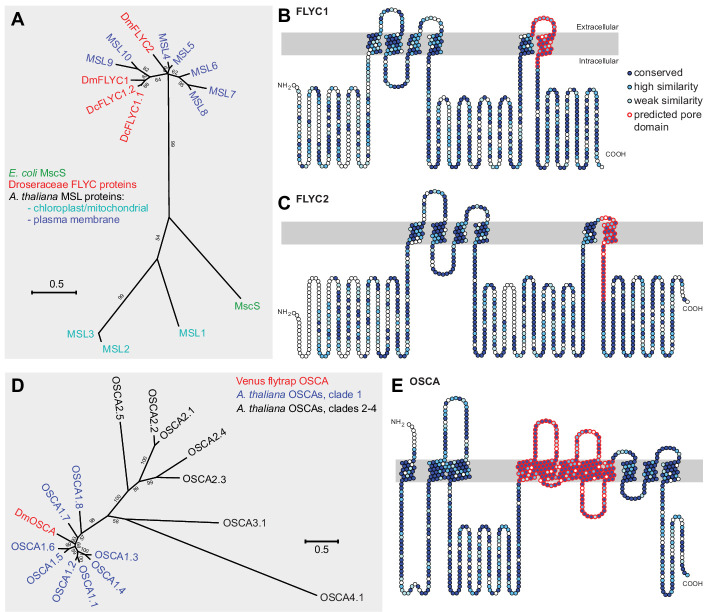
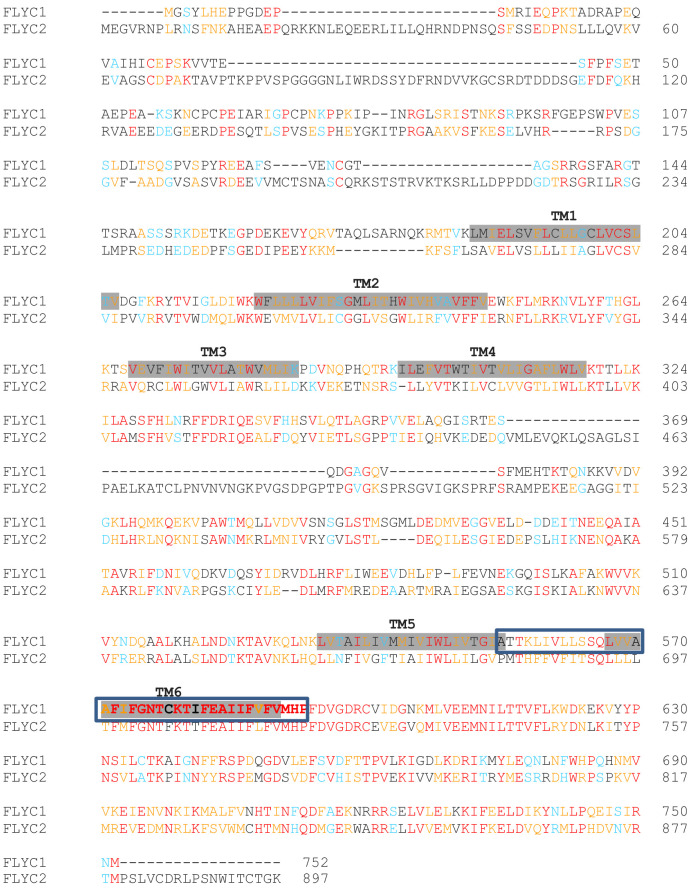
Figure 2. Molecular phylogenetic relationship of FLYCATCHER and OSCA proteins.
(A) Phylogenetic analysis by maximum likelihood method to show the relationship between the conserved MscS domain (Haswell and Meyerowitz, 2006) of Escherichia coli MscS protein, Arabidopsis thaliana MSL proteins (MSL1–MSL10), and Dionaea muscipula/Venus flytrap (DmFLYC1 and DmFLYC2) and Drosera capensis/Cape sundew (DcFLYC1.1 and DcFLYC1.2) FLYCATCHER proteins. (B, C) Predicted topology for DmFLYC1 (B) and DmFLYC2 (C) proteins. (D) Phylogenetic analysis by maximum likelihood method showing the relationship between A. thaliana OSCA family proteins and Venus flytrap DmOSCA. (E) Predicted topology for DmOSCA protein. In (A) and (D), bootstrap values >50 are shown; scale, substitutions per site. In (B), (C), and (E), amino acid residues that are conserved with Arabidopsis MSL10, MSL5, and OSCA1.5, respectively, are indicated in dark blue circles, whereas residues similar in identity are indicated in lighter blue circles. The predicted pore domain for each protein is indicated in red circles.