Fig. 1 |. NKG7 is highly upregulated in splenic CD4+ T cells during L. donovani infection.
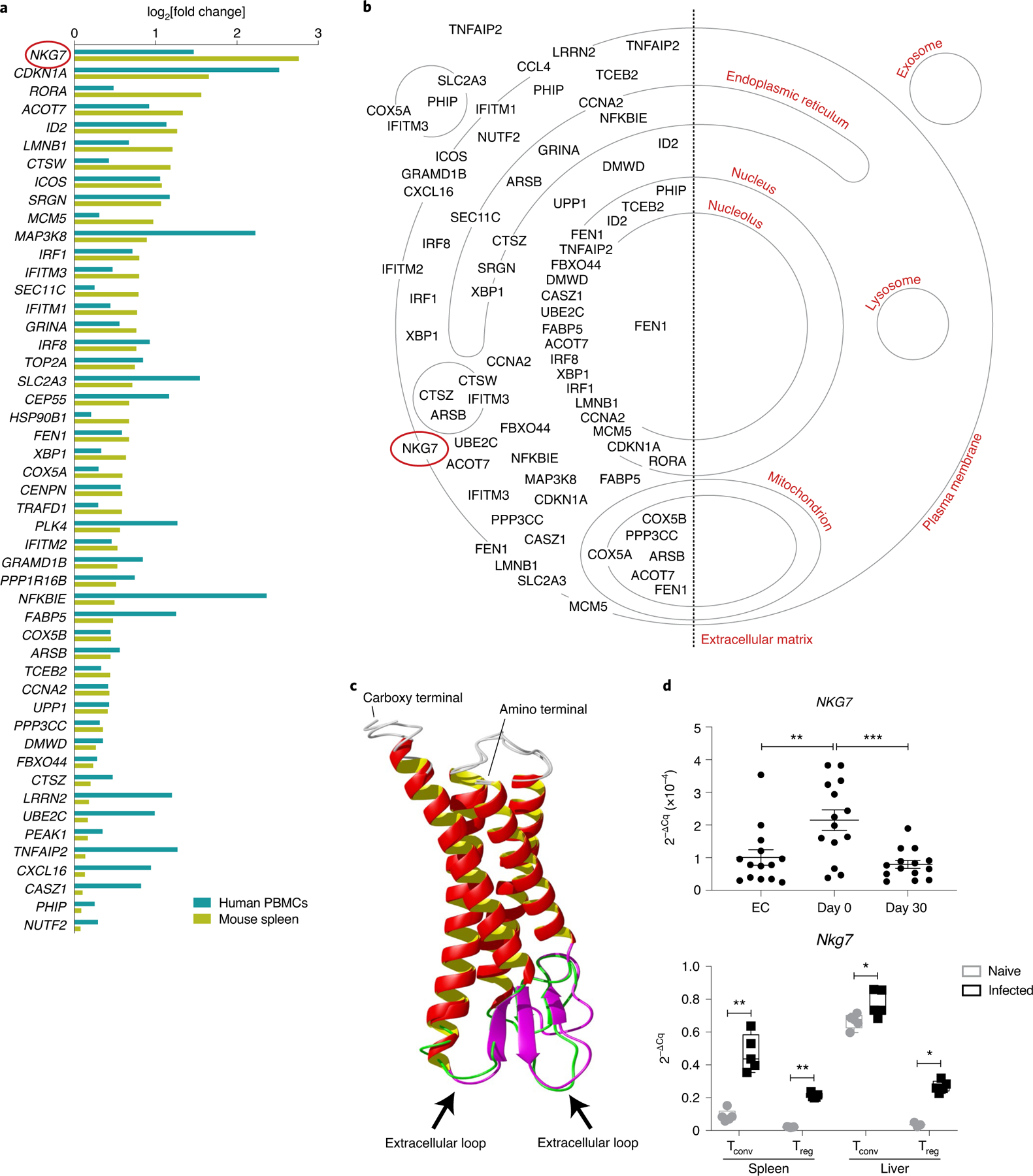
a, Upregulated genes found in CD4+ T cells isolated from mouse spleen at day 56 p.i. and in human PBMCs from patients with visceral leishmaniasis at the time of admission to clinic for treatment. b, Cellular locations of proteins encoded by the upregulated genes in a are indicated on the cellular map using information obtained from the Gene Ontology Cellular Component knowledge base. c, The protein structure of NKG7 generated using I-TASSER. Predicted extracellular loops (indicated by black arrows) of human and mouse NKG7 are highlighted in green and purple, respectively. d, Top: validation of NKG7 upregulation in patients with visceral leishmaniasis before treatment (day 0; n = 14) compared with the same patients after treatment (day 30) and endemic controls (EC; n = 14) by RT-qPCR. Bottom: RT-qPCR validation was also performed in conventional T cells (Tconv) and Treg cells from the spleen and liver of naive and infected (day 56 p.i.) mice. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons test (top) or two-way ANOVA with Šídák’s multiple comparisons test (bottom). Center lines indicate median values, box limits indicate upper and lower quartiles, and whiskers indicate maximum and minimum measures. *P < 0.05; **P < 0.01; ***P < 0.001.
