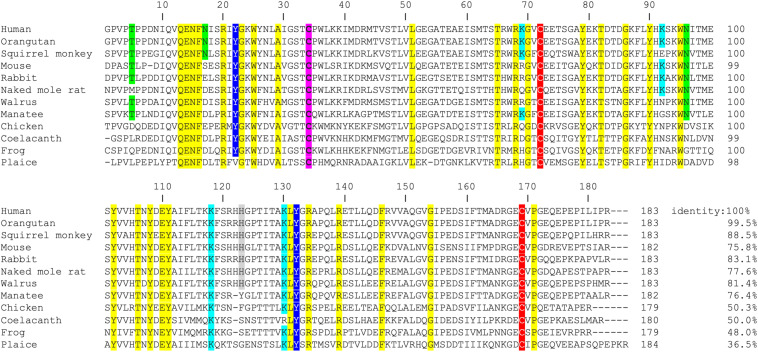
FIGURE 2.
Amino acid sequence alignment of A1M from 12 different species. The amino acid sequence of human wild-type (wt) A1M and 11 additional species were retrieved from public data bases (www.ncbi.nlm.nih.gov and www.uniprot.org) and aligned using the http://www.ebi.ac.uk/Tools/msa/clustalw2/ software. The degree of identity of the different sequences to the human sequence is presented as percent relative to human A1M. Amino acids with reported functional impact in human A1M are marked: the C34, described to be important for reduction and antioxidant properties as well as heme binding, is marked with pink. Glycosylated positions (T5, N17, and N96) are marked with green. Lysine residues (K69, K92, K118, and K130), described to be related to the yellow-brown modifications, are marked with light blue. The disulfide bridge, formed between C72 and C169, is marked in red. H123, suggested to take part in coordination of heme-iron, is marked with gray. Tyrosines (Y22 and Y132), shown to be modified in radical scavenging reactions are marked in dark blue. Of these functionally important amino acids, five are completely conserved in all 12 species (C34, C72, K118, Y132, and C169). Additional 28 amino acids, which are identical between all species in the set, are marked with yellow. Human, Homo sapiens; Orangutan, Pongo abelii; Squirrel monkey, Saimiri boliviensis boliviensis; Mouse, Mus musculus; Rabbit, Oryctolagus cuniculus; Naked mole rat, Heterocephalus glaber; Walrus, Odobenus rosmarus divergens; Manatee, Trichechus manatus latirostris; Chicken, Gallus gallus; Coelacanth, Latimeria chalumnae; Frog, Xenopus laevis; and Plaice, Pleuronectes platessa.