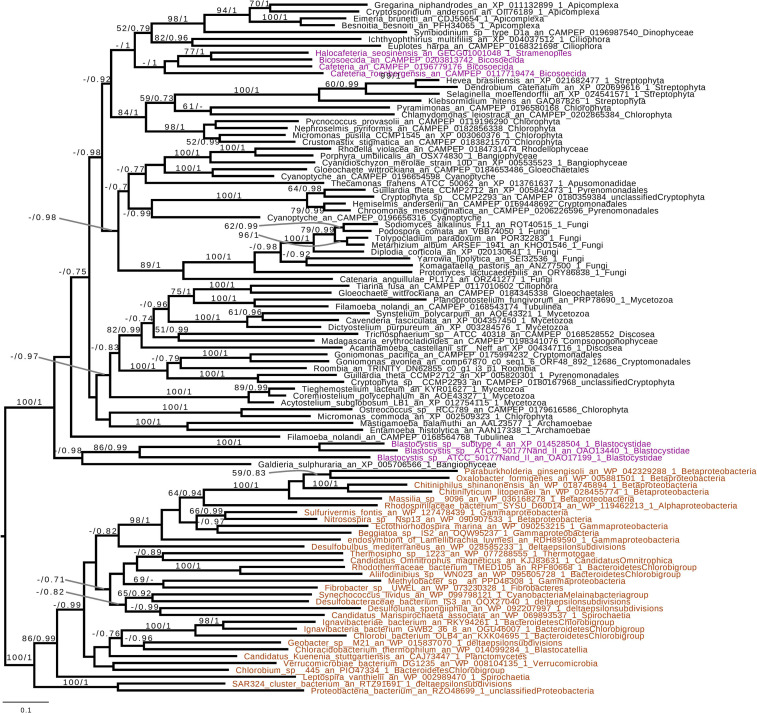
FIGURE 3.
Phylogenetic analysis of glycogen phosphorylase (GT35 family). The tree displayed is midpoint rooted and represents the consensus tree obtained with Phylobayes 4.1 with ML bootstrap values drawn from 100 bootstraps repetition with IQTREE (left) and Bayesian posterior probabilities (right) mapped onto the nodes. Bootstrap values >50% are shown, while only posterior probabilities >0.6 are shown. The scale bar shows the inferred number of amino acid substitutions per site. Sequences are highlighted in purple for Stramenopila, brown for Bacteria, while everything else is in black. Sequence names are composed of the organism name, the accession number, and their clades. We can see that Bicosoecida, Halocafeteria, and Cafeteria group together, with a high posterior probability (pp = 1). However, Blastocystis was not found inside a monophyletic group with Stramenopila but was found among other eukaryotes. The Blastocystis position is probably due to the fast-evolving sequence in this organism as it has been observed in several studies (Eme et al., 2017; Moreira and López-García, 2017).