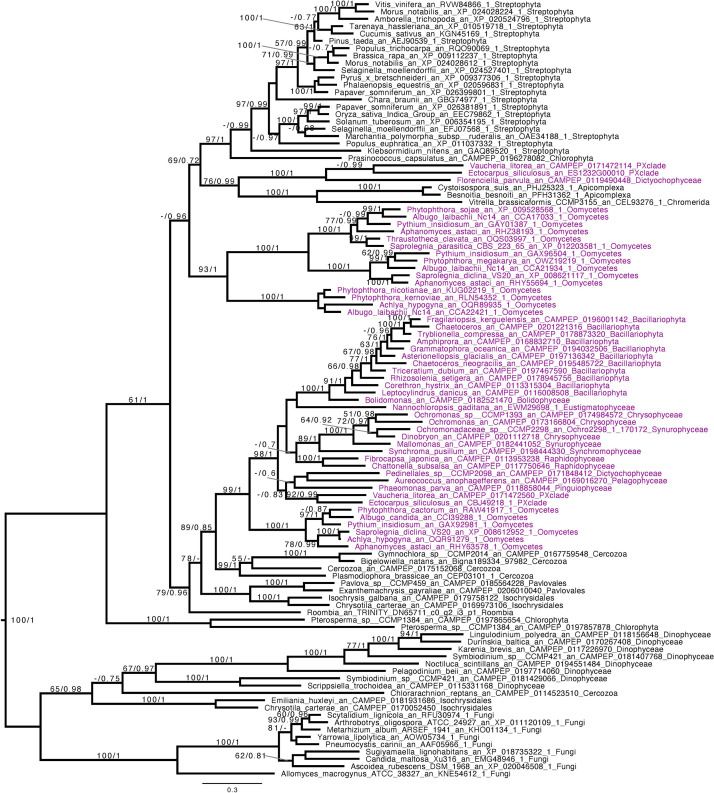
FIGURE 4.
Phylogenetic analysis of GT48. The tree displayed is midpoint rooted and represents the consensus tree obtained with Phylobayes 4.1 with ML bootstrap values drawn from 100 bootstraps repetition with IQTREE (left) and Bayesian posterior probabilities (right) mapped onto the nodes. Bootstrap values >50% are shown, while only posterior probabilities >0.6 are shown. The scale bar shows the inferred number of amino acid substitutions per site. Sequences are highlighted in purple for Stramenopila, while everything else is in black. Sequence names are composed of the organism name, the accession number, and their clades. We can observe a strongly supported group with all Gyrista (BS = 99, pp = 1), with inside the expected topology based on Stramenopila phylogeny. In addition, among those sequences we find the characterized enzyme from P. tricornutum. Then, this group of sequence is likely to be the one involved in laminarin biosynthesis. Moreover, we can find close to this Gyrista group several sequences from both Haptophyta and Cercozoa (BS = 79, pp = 0.96), probably also involved in storage polysaccharide metabolism.