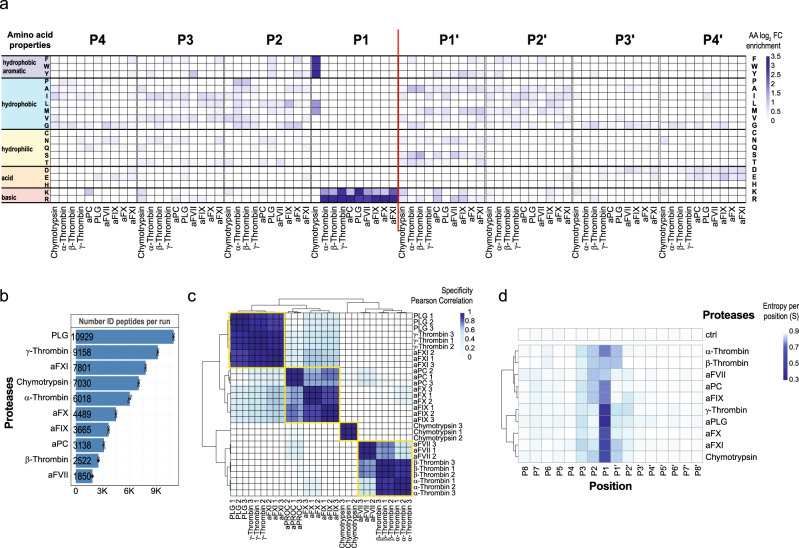
Fig. 3. High-throughput screening of coagulation proteases.
a Positional substrate preferences of coagulation proteases from the chymotrypsin-like family (Chymotrypsin, activated α-, β-, γ-Thrombin, aFVII, aFIX, aFX, aFXI, aPC and PLG). The heat map includes positions P4-P4’. The AAs are grouped according to their physicochemical properties. The enrichments are reported as log2 FC compared to a random AA distribution generated from HTPS database. b Peptides generated by the coagulation proteases included in the study; each protease is characterized by the average number of peptides identified from three independent replicate experiments (n = 3). Data are presented as mean values±SD. c Unsupervised hierarchical cluster of coagulation proteases. The color scale describes the Pearson correlation coefficient value calculated for the respective protease samples. d Unsupervised hierarchical cluster of coagulation proteases according to the positional cleavage entropy. The color scale describes the cleavage entropy (S) values for positions P8-P8’ for all respective proteases included in the assay. Source data are provided as a Source Data file.