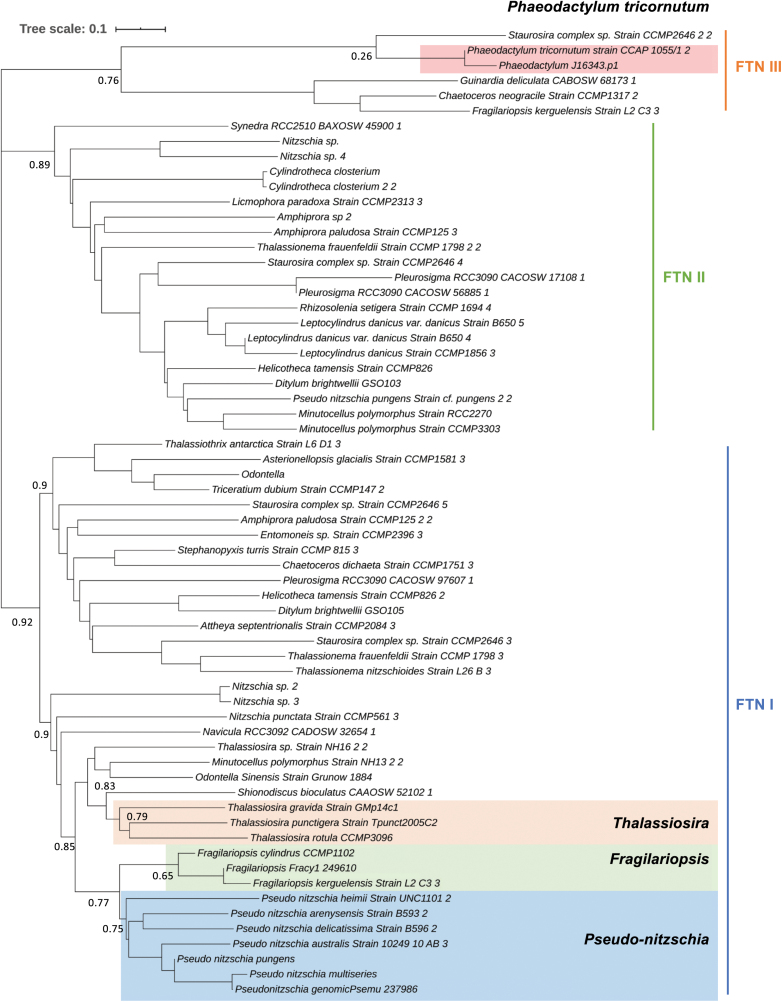
Fig. 6.
Phylogenetic tree of ferritins (FTNs) in diatoms. The phylogenetic tree was constructed to clarify the evolutionary distance of FTN between species. A total of 244 sequences were first retrieved by a HMMER search using the FTN PFAM domain PF00210 as a query in the available 82 diatom transcriptomes/genomes with an E-value cut-off of 1×10–10. Retrieved sequences were analysed using the CD-HIT web server and sequences that met a similarity threshold >0.9 were presumed to be duplicates and eliminated. We generated a Hidden Markov Model for FTN using P. tricornutum and closely aligned sequences retrieved by BLASTp, and this was used as the basis for a second HMMER search of the remaining sequences, with an E-value cut-off of 1×10–10, to further reduce the redundancy. This left a total of 64 representative sequences, which captured the diversity of FTNs within the diatoms. Conserved sequences were aligned using the alignment builder in Geneious v.10.2 under default criteria. The tree was drawn with ITOL (https://itol.embl.de/). Numbers beside branches indicate RaxML bootstrap coefficients.