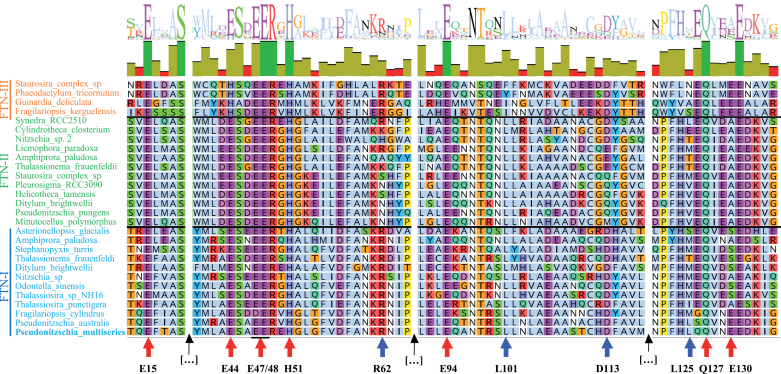
Fig. 7.
Protein sequence alignment of representative diatom ferritins (FTNs). FTN protein sequence alignment (one-letter code) was carried out using MUSCLE in Geneious v.10.2. At the top is the sequence logo and identity. At the bottom, red arrows highlight the ferroxidase residue sites first identified in Pseudo-nitzschia multiseries (Pfaffen et al., 2013). The glutamic acid residues (E15, E47/48, and E94), one glutamine residue (Q127), and the histidine residue H51 are conserved across the three different clades (red arrows). However, the glutamic acid residue E44, which is conserved in FTN I/II, is replaced with histidine in FTN III. These residues are involved in iron binding. Blue arrows show conserved residue pairs essential for iron release in the frog species Rana catesbeiana (Jin et al., 2001). The amino acid pairs R62/D113 and L101/L125 are adjacent in three-dimensional space. R62/D113 is conserved in both FTN I and FTN III but not FTN II (which shows significant variation in these positions). Leucine is conserved in position 101 in FTN I and II but not FTN III, while leucine at position 125 is not conserved in any diatoms apart from two Pseudo-nitzschia species and Synedra sp. RCC2510. The most common replacement for leucine in this position is methionine, as observed in F. cylindrus and Thalassiosira sp. NH16. Black arrows indicate breaks in the sequence.