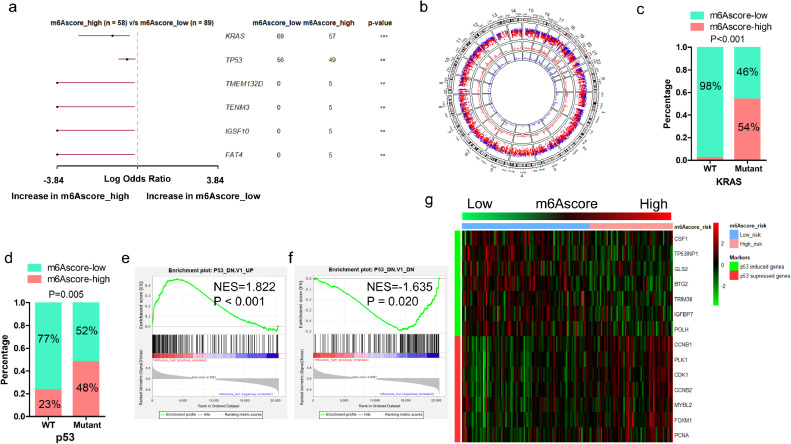
Fig. 6.
Comparison of mutation landscape between m6Ascore-high and m6Ascore-low tumors. (a) Forest plot shows the most significantly mutant genes that vary between m6Ascore-high and m6Ascore-low tumors (**P < 0.01; ***P < 0.001); (b) A circular plot shows the mutation profile of m6Ascore-high and m6Ascore-low tumors. From the most inner to the most outer circles listed the genomic density of mutation genes in m6Ascore-high tumors, in m6Ascore-low tumors, and genomic rainfall of mutation genes in all samples; (c) Proportion of m6Ascore-high and m6Ascore-low group in patients harbored wildtype or mutant KRAS; (d) Proportion of m6Ascore-high and m6Ascore-low group in patients harbored wildtype or mutant TP53; (e) GSEA analysis of genes up-regulated in NCI-60 panel of cell lines with mutated TP53; (f) GSEA analysis of genes down-regulated in NCI-60 panel of cell lines with mutated TP53; (g) Correlation of m6Ascore and the expression of p53-induced genes or p53-suppressed genes in PDAC patients.