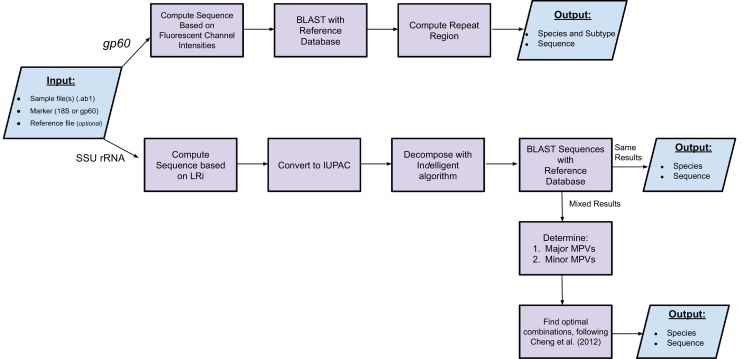
Fig. 1.
CryptoGenotyper schematic workflow.
The program begins with the user inputting the gene target, sample chromatograms and database (optional). (a) If chromatograms correspond to the gp60 gene target, the sequence is retrieved by analyzing the fluorescent channel intensities. A homology search is performed against the reference database and the repeat region is calculated. (b) If chromatograms correspond to the SSU rRNA gene, the sequence is computed based on the log ratio of intensity and converted to IUPAC based nucleotide code where double peaks appear. Afterwards, the sequence is decomposed with Indelligent (Dmitriev and Rakitov, 2008) and a homology search is performed using BLAST against the reference database. If mixed sequences are determined, they are classified by following the protocol outlined in Chang et al. (2012) for determining the most possible variances (MPVs) and optimal combinations. For both markers, the sequence and species and/or subtype information is outputted.