Abstract
Moricandia is a genus belonging to the family Brassicaceae. C3 and C3—C4 photosynthesis Moricandia species exist in a close phylogenetic proximity, as well as to Brassica crops. Here, we performed PacBio genome sequencing on M. moricandioides and M. arvensis. The genomes were assembled using Flye assembler, then polished with Illumina reads and reduced duplication with Purge Haplotigs. The total length of genome assemblies of M. moricandioides and M. arvensis was 498 Mbp and 759 Mbp, respectively. These data will be useful for studies of the genetic control of C3—C4 characteristics, therefore gaining new insights into the early evolutionary steps of C4 photosynthesis. Further, it can be integrated into Brassica crop breeding. The data can be accessed at ENA under the project number PRJEB39764.
Keywords: Moricandia, PacBio sequencing, NGS sequencing, Genome assembly
Specifications Table
| Subject | (Plant) Biology |
| Specific subject area | Genomics |
| Type of data | Table, figure, genomic assembly data in FASTA format |
| How data were acquired | Whole genome sequencing was performed using the PacBio RSII platform |
| Data format | Raw and analyzed |
| Parameters for data collection | gDNA was extracted from mature leaf tissues of M. moricandioides and M. arvensis |
| Description of data collection | Sequencing reads were generated on the PacBio RS II platform. The reads were then assembled de novo using Flye assembler. |
| Data source location | Düsseldorf, Germany |
| Data accessibility | Accessible as a project on European Nucleotide Archive (PRJEB39764) https://www.ebi.ac.uk/ena/submit/sra/#studies Link for direct download of raw data: <https://www.ebi.ac.uk/ena/browser/view/PRJEB39764> Link for direct download of FASTA-format assemblies: <https://www.ebi.ac.uk/ena/browser/view/GCA_905132765.1> <https://www.ebi.ac.uk/ena/browser/view/GCA_905132885.1> |
Value of the Data
-
•
M. moricandioides and M. arvensis are closely related sister species within the crucifers (Brassicaceae)
-
•
M. moricandioides performs C3 photosynthesis whereas M. arvensis performs C3—C4 intermediate photosynthesis
-
•
These are the first whole genome assemblies of M. moricandioides and M. arvensis
-
•
The genome assemblies provide the basis for identification of genetic factors underpinning C3—C4 photosynthesis, through comparative genomics
-
•
C3—C4 photosynthesis is an intermediary step on the evolutionary trajectory from C3 to C4 photosynthesis; hence the data will provide novel insights into the early evolutionary steps into the direction of C4 photosynthesis.
-
•
The genome data serve as a valuable resource for engineering of the photosynthetic efficiency of C3 plants.
1. Data Description
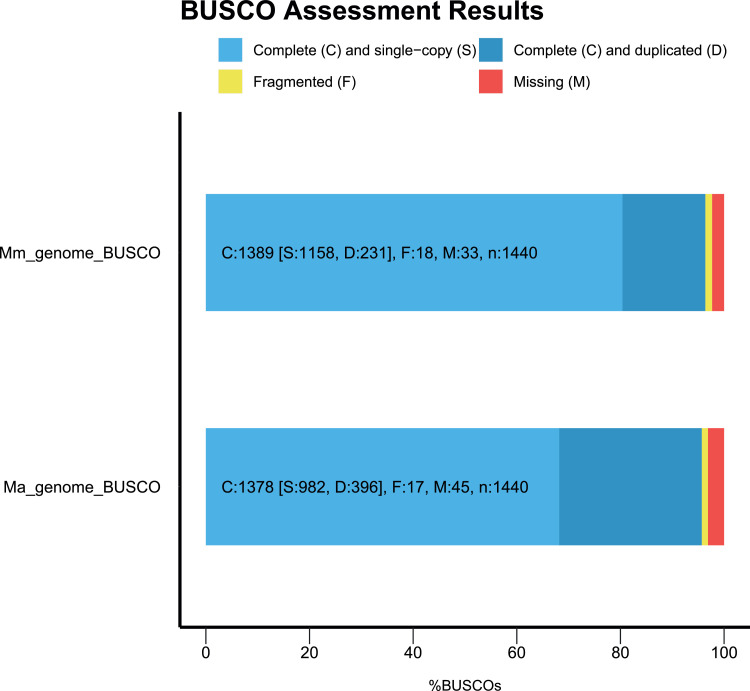
We present here the genome sequencing and assembly of C3 M. moricandioides and C3—C4 M. arvensis. A total output of 43 and 71.9 Gb was generated by PacBio sequencing for M. moricandioides and M. arvensis, respectively. The de novo assembly was performed using Flye assembler, followed by reducing the duplication with Illumina reads through Purge Haplotigs. The resulting draft genome of M. moricandioides was 498,312,484 bp in size with a GC content of 36.02% with N50 contig length of 114.3 kb. And the assembled genome size of M. arvensis was 758,710,152 bp with a GC content of 36.75% with N50 contig length of 82.1 kb. Main de novo assembly statistics of the Moricandia genome assemblies are shown in Table 1. We evaluated the assembled genomes by Benchmarking Universal Single-Copy Orthologs (BUSCO) (Fig. 1). The majority (96.4% and 95.7% of M. moricandioides and M. arvensis, respectively) was complete BUSCO genes. For the rest of the genomes, the amount of fragmented genes was 1.3% and 1.2% and the missing genes presented 2.3% and 3.1% for M. moricandioides and M. arvensis, respectively. The genome assembly of M. moricandioides comprised more complete and single copy BUSCOs (83.4% of the complete BUSCOs) than M. arvensis genome assembly (71.3% of the complete BUSCOs).
Table 1.
Main de novo assembly statistics.
| M. moricandioides | M. arvensis | |
|---|---|---|
| Yield | 43 Gb | 71.9 Gb |
| Mean read length | 7.5 kb | 10.2 kb |
| SMRT cells | 50 | 56 |
| Yield/SMRT cell | 0.859 Gb | 1.284 Gb |
| Genome size estimated (Gb) | 0.66 Gb | 0.737 Gb |
| Coverage (est. genome size) | 65 | 98 |
| Coverage (assembled genome size) | 86 | 95 |
| Corrected bases (Gb) | 21.2 | 40.8 |
| Trimmed bases (Gb) | 21.2 | 40.2 |
| Bases assembled (bp) | 498,312,484 | 758,710,152 |
| No. contigs | 9638 | 15,876 |
| N50 (kb) | 114.3 | 82.1 |
| GC content | 36.02% | 36.75% |
Fig. 1.
Genome assembly assessment by BUSCO.
BUSCO completeness assessments of M. moricandioides (Mm) and M. arvensis (Ma) genome assembly. The proportions of complete (C, sum of light and dark blue bars), complete single-copy (S, light blue), complete duplicated (D, dark blue), fragmented (F, yellow), and missing (M, red) BUSCOs are shown in the bar chart.
2. Experimental Design, Materials and Methods
2.1. Sampling and DNA extraction
For PacBio sequencing, plants of M. moricandioides (Botanical Garden Osnabrück: 04–0393–10–00) and M. arvensis (IPK Gatersleben: MOR1) were grown in soil under controlled conditions in the CLF climate chamber under conditions of 12 h day (23 °C, ca. 200 µmol m-2 s-1)/12 h night (20 °C) in December 2015; plants of M. arvensis (IPK Gatersleben: MOR1) were grown under the same conditions in June 2016. For Illumina sequencing, plants of M. moricandioides and M. arvensis were grown under the same conditions in April/May 2019. The whole plant shoot samples were harvested ca. five weeks after germination and stored at −80 °C. For preparation of gDNA, nuclei were extracted first after the method described by [1]. Nuclei were treated with RNAse A (10 mg/ml) for 30 min at 37 °C followed by incubation with protease K (0.8 mg/ml) for 2 h at 50 °C. The DNA samples were purified using Qiagen G20 tips (Qiagen, Germany) following manufacturer's instructions. DNA was resuspended in TE (10 mM Tris, pH 8.0, 1 mM EDTA).
2.2. Library preparation and sequencing
The preparation of the M. moricandioides sequencing library was made by the SMRTbell Express Template Prep Kit 3.0 and whole genome sequencing was performed on PacBio RS II platform (Pacific Biosciences, Menlo Park, CA, USA) by the Max Planck-Genome-centre Cologne, Germany (https://mpgc.mpipz.mpg.de/home/). The M. arvensis gDNA was constructed into a library using the SMRTbell Express Template Prep Kit 2.0 and was sequenced on PacBio RS II platform by KeyGene, N. V., Wageningen, Netherlands (https://keygene.com). In total, the output of 43 and 71.9 Gb was generated for M. moricandioides and M. arvensis, respectively. These yielded at least 86 × coverage of each genome.
The gDNA of M. moricandioides and M. arvensis was subjected to a paired-end library preparation for genome sequencing on the Illumina HiSeq 3000 platform (Illumina, San Diego, USA) by the Biologisch-Medizinisches Forschungszentrum (BMFZ) of the Heinrich-Heine University (Düsseldorf, Germany). Prior to library preparation 650 ng of gDNA were sheared with Covaris ME220 (Covaris, Inc.) to a mean fragment size of 350 bp. Library preparation was performed according to the manufacturer's protocol by using the VAHTS Universal DNA Library Prep Kit for Illumina (Vazyme Biotech Co.; Ltd) without any amplification step and an additional size selection step after adaptor ligation to get rid of smaller fragments. Library was quantified via qPCR by using KAPA library quantification kit (Roche Diagnostics Corporation) on a QuantStudio 3 (Thermo Fisher Scientific Inc.) and subsequently sequenced on a HiSeq3000 system (Illumina Inc) with a read setup of 2 × 151 bp. 114 and 167 million reads per library were obtained from M. moricandioides and M. arvensis, respectively.
2.3. Quality control, genome assembly, and genome assessment
De novo assembly of the PacBio reads was performed by Flye [2] with the estimated genome size set to 1Gbp for both species. The PacBio reads were mapped back to the assembly with BLASR [3] via pbalign (https://github.com/PacificBiosciences/pbalign), and used to generate a polished consensus with Arrow (https://github.com/PacificBiosciences/GenomicConsensus). The Illumina reads were quality controlled using FASTQC, mapped using BWA [4], and used to polish the genome assembly through Pilon [5]. Finally the PacBio reads were mapped to the twice-polished assembly using Minimap2 with PacBio preset settings [6], and then used for the reduction of duplication with Purge Haplotigs [7]. Coverage thresholds of 6, 42, 140 (M. moricandioides) and 4, 32, 140 (M. arvensis) for Purge Haplotigs’ low, middle, and high coverage, respectively. The genome assembly evaluation was conducted by BUSCO v3 [8] and QUAST [9].
CRediT Author Statement
Nils Koppers: Conceptualization, Methodology, and Software; Meng-Ying Lin: Data curation, Writing - Original draft preparation, and Visualization; Alisandra Denton: Data curation and Validation, Writing - Reviewing and Editing; Urte Schlüter: Sample preparation, Reviewing and Editing; Andreas P.M. Weber: Supervision, Reviewing and Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgements
We thank the Max Planck-Genome-Centre Cologne (http://mpgc.mpipz.mpg.de/home/) for performing library preparation and whole genome sequencing of M. moricandioides on PacBio RSII platform. For the library preparation and whole genome sequencing of M. arvensis using PacBio RSII platform, we appreciate the support from the keyGene, N.V. (http://keygene.com). We also thank the BMFZ (Biologisch-Medizinisches Forschungszentrum) of the Heinrich-Heine University (Düsseldorf, Germany) for preparing gDNA library and performing whole genome sequencing on Illumina platform. We are thankful for the computational resources provided by the HHU Centre for Information and Media Technology (ZIM). This work was funded by grants of the Deutsche Forschungsgemeinschaft to APMW under Germany's Excellence Strategy EXC-2048/1, Project ID 390686111, and by ERA-CAPS project C4BREED (WE 2231/20–1), and by graduate fellowships of the International Max Planck Research School on "Understanding Complex Plant Traits using Computational and Evolutionary Approaches" to MY.L and of the DFG IRTG 1525 iGRADplant to N.K.
References
- 1.Zhang M., Zhang Y., Scheuring C.F., Wu C.-.C., Dong J.J., Zhang H.-.B. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat. Protoc. 2012;7(3):467–478. doi: 10.1038/nprot.2011.455. [DOI] [PubMed] [Google Scholar]
- 2.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 3.Chaisson M.J., Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13(1):238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio] 2013 http://arxiv.org/abs/1303.3997 Accessed November 17, 2020. [Online]. Available: [Google Scholar]
- 5.Walker B.J. Pilon: an Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roach M.J., Schmidt S.A., Borneman A.R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]