Figure 8.
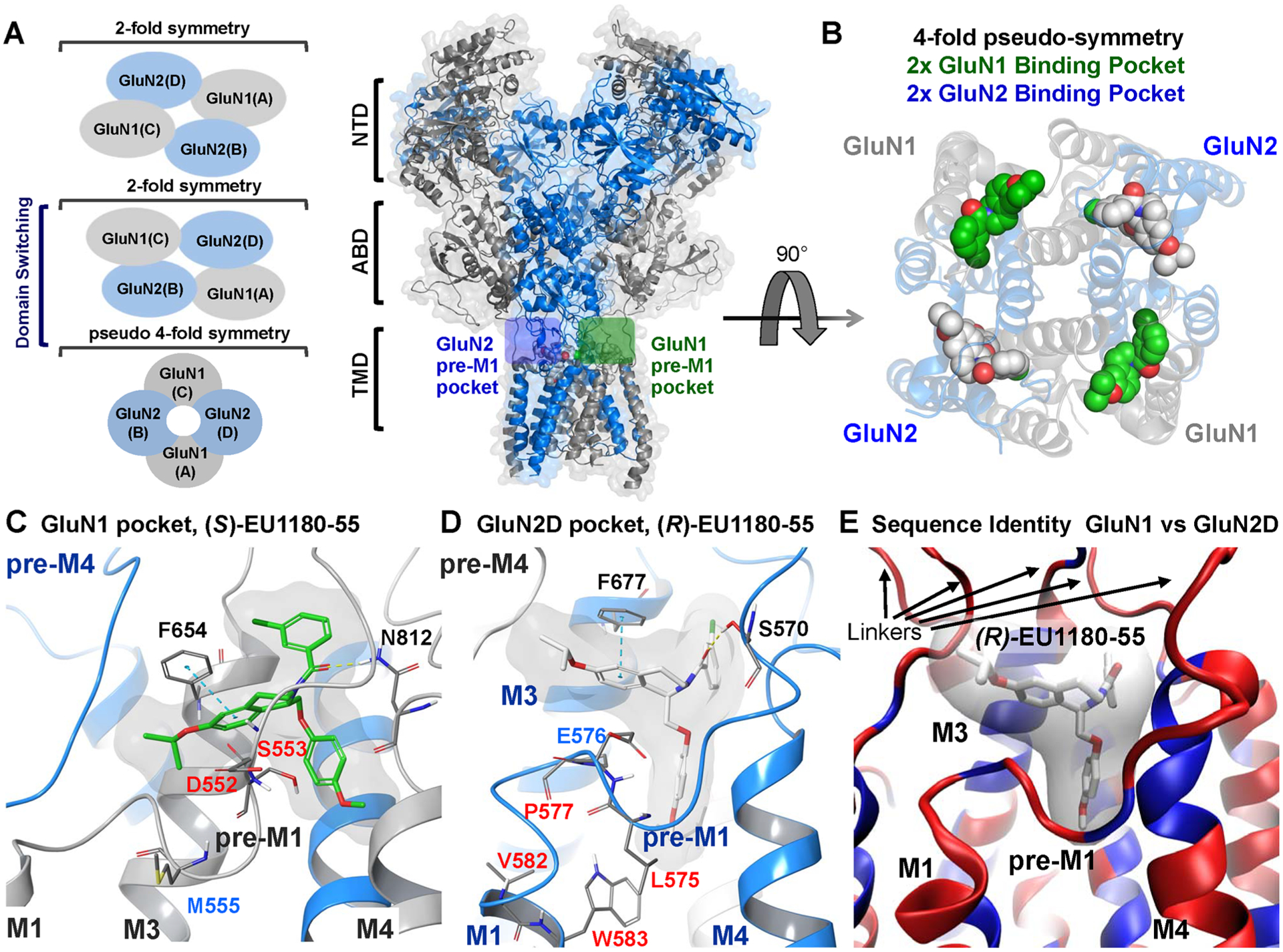
(A) Cartoon representation of a GluN1/GluN2D NMDAR homology model bound with EU1180–55 enantiomers in the GluN1 (green box) and GluN2 (blue box) pre-M1 pockets. Blue represents the GluN2 subunits and gray the GluN1 subunits. (left) Representation of the fold symmetry in the NMDAR of the NTD, ABD, and TMD. (B) Rotation (90°) and removal of the ABD and NTD reveals the pseudo-4-fold symmetry of the transmembrane region with the (S) and (R) enantiomers of EU1180–55 bound. Green shows the (S)-EU1180–55 enantiomer, which mutagenesis suggests binds to a pocket on GluN1. White shows (R)-EU1180–55 bound to the analogous pocket on GluN2. (C, D) Expanded view of (S)-EU1180–55 (green) within the GluN1 binding pocket and (R)-EU1180–55 (white) bound within a pocket on GluN2D. If residues were tested in either Figure 6 or 7, the residues numbers are colored using the color scheme in Figures 6 and 7. (E) Overlay of the GluN1 and GluN2D binding pockets with blue showing identical residues and red showing nonidentical residues; only a single ribbon is shown. The binding pocket is conserved with most of the variation within pre-M1 and the linker region between the TMD and the ABD. See Supplemental Figure S8 for more information.
