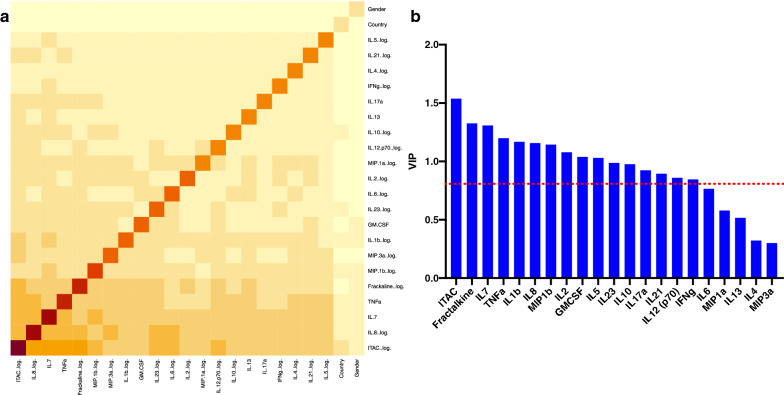
Fig. 2.
Partial least square (PLS) and Random Forest Analysis supported initial analyses. a Partial Least Square (PLS) Analysis for combined Zambian and Rwandan cohorts. Analysis was done with NIPALS Fit with 1 Factor. VIP (Variable Importance Plot) Threshold was set at 0.8. ITAC, GMCSF, Fractalkine, IFNg, IL-10, IL-12, IL-17a, IL-1b, IL-2, IL-21, IL-23, IL-5, IL-7, IL-8, MIP-1b, and TNFa were the biomarkers that caused preinfection profile in combined cohorts. b Random Forest Analysis used to determine which cytokines and chemokines contributed to the pattern of predisposition observed. In this analysis, the color of the square indicates the importance of the cytokine or chemokine to the profile of predisposition (light yellow is less importance, dark red is high importance)