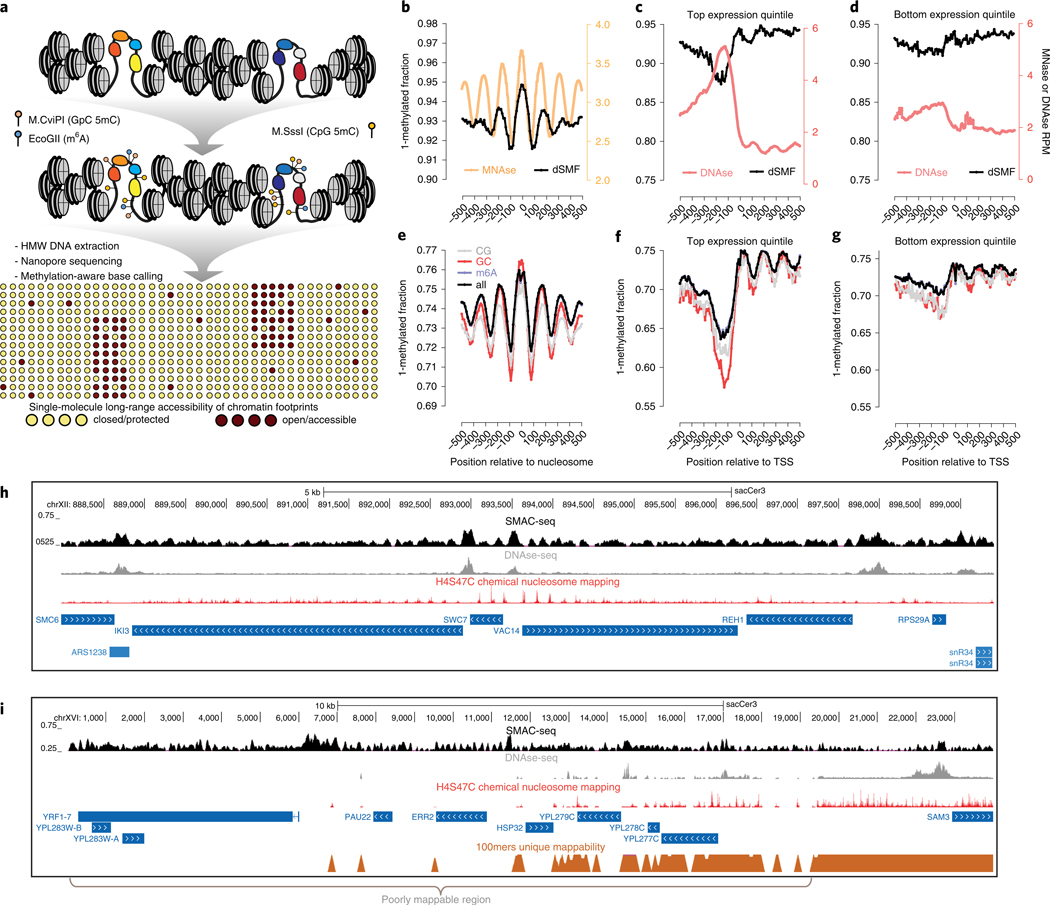
Fig. 1 |. The SMAC-seq assay for profiling chromatin accessibility and nucleosome positioning at the multikilobase scale.
a, Outline of the SMAC-seq assay. Intact chromatin is treated with m6A and CpG and GpC 5mC methyltransferases, which preferentially methylate DNA bases in open chromatin regions. High molecular weight (HMW) DNA is then isolated and subjected to nanopore sequencing, and methylated bases are used to reconstruct the open chromatin state within individual molecules. b-h, SMAC-seq faithfully captures chromatin accessibility around promoters and positioned nucleosomes in S. cerevisiae. b, MNAse-seq and dSMF profiles around chemically mapped positioned nucleosome dyads. c, DNAse-seq and dSMF profiles around the top 20% highly expressed genes in S. cerevisiae. d, DNAse-seq and dSMF profiles around the bottom 20% expressed genes in S. cerevisiae. RPM, reads per million (c,d). e, Average SMAC-seq profile around chemically mapped positioned nucleosomes dyads (shown is the ‘diamide 0 min rep2’ sample). f, Average SMAC-seq profile around the top 20% highly expressed genes in S. cerevisiae. g, Average SMAC-seq profile around the bottom 20% expressed genes in S. cerevisiae. TSS, transcription start site (f,g). h, SMAC-seq correlates closely with both DNAse-seq and nucleosome occupancy profiling at the level of individual loci and provides a combined readout of accessibility and nucleosome positioning. Shown is the aggregate SMAC-seq signal along the genome (aggregated over 50-bp windows sliding every 5 bp; see Methods for details), together with DNAse-seq, nucleosome chemical mapping data and transcriptional activity (measured by PRO-seq and PRO-cap). Large aggregate SMAC-seq signal enrichments match closely with DNAse accessibility peaks, while smaller aggregate SMAC-seq peaks are inversely correlated with positioned nucleosomes. i, SMAC-seq profiles chromatin accessibility in repetitive regions of the genome that are ‘invisible’ to short reads. The telomeric region of chrXVI is shown.