Mechanisms of myogenic commitment are deciphered through omics analysis of hiPSC-specific lineage specification measurements.
Abstract
Understanding the mechanisms of myogenesis in human induced pluripotent stem cells (hiPSCs) is a prerequisite to achieving patient-specific therapy for diseases of skeletal muscle. hiPSCs of different origin show distinctive kinetics and ability to differentiate into myocytes. To address the unique cellular and temporal context of hiPSC differentiation, we perform a longitudinal comparison of the transcriptomic profiles of three hiPSC lines that display differential myogenic specification, one robust and two blunted. We detail temporal differences in mechanisms that lead to robust myogenic specification. We show gene expression signatures of putative cell subpopulations and extracellular matrix components that may support myogenesis. Furthermore, we show that targeted knockdown of ZIC3 at the outset of differentiation leads to improved myogenic specification in blunted hiPSC lines. Our study suggests that β-catenin transcriptional cofactors mediate cross-talk between multiple cellular processes and exogenous cues to facilitate specification of hiPSCs to mesoderm lineage, leading to robust myogenesis.
INTRODUCTION
Skeletal muscle progenitors derived from human induced pluripotent stem cells (hiPSCs) have the potential to play a central role in the realization of individualized therapies (1). Chief among these unmet clinical needs are in vitro patient-specific tissue models for pharmacologic studies, as well as cell-based therapies for age- or injury-related muscle loss, neuromuscular disorders, and muscular dystrophies (2, 3). hiPSCs offer unique benefits: They give rise to cells from all three germ layers, are autologous, and have indefinite in vitro expansion (4). However, cell line–to–cell line variability in robust and reliable myogenic differentiation of hiPSCs remains a major bottleneck (5). A first step to address this challenge is a better understanding of the multiple intra- and extracellular mechanisms that underlie myogenic differentiation in the specific context of hiPSCs.
Studies of precommitted progenitors in vitro and embryonic development in vivo illustrate the context-dependent and multifactorial nature of myogenesis. Promyogenic medium conditions that lead to in vitro myogenic maturation were originally developed in studies of precommitted progenitors (3, 6). In addition to soluble factors, cell-matrix and cell-cell interactions also play a key role in myogenic specification (7). Embryological studies of myogenesis reveal finely tuned spatiotemporal regulation, including paracrine gradients that modulate signaling pathways as well as an internal “segmentation clock,” that guides paraxial mesoderm development and subsequent somitogenesis and limb bud formation (3). In vitro and in vivo studies delineate the crucial roles of regulatory transcription factors, such as MYOD and MYOG, in myogenic specification, as well as PAX7, a marker for satellite cells, the endogenous stem cells of skeletal muscle (2, 8, 9).
Methods for inducing myogenic differentiation in human pluripotent stem cells (hPSCs) leverage knowledge of in vitro myogenesis of precommitted progenitors and in vivo development. Embryoid body (EB)–based protocols attempt to recapitulate embryonic-like conditions in suspension culture to induce spontaneous differentiation of all three germ layers; mesodermal progenitors are then selected and matured in promyogenic culture conditions (10). However, due to unpredictable spontaneous differentiation of EBs, monolayer culture has emerged as the method of choice for myogenic induction of hiPSCs. Within monolayer culture, there are two main strategies: ectopic overexpression of a myogenic regulatory factor, such as MYOD or PAX7, or directed differentiation with soluble cues, or combination thereof (3, 9, 11–13). The transgene-free directed differentiation methods attempt to mimic signaling pathway modulation of embryonic myogenesis by using sequential, defined cocktails of small molecules and growth factors (11, 12).
However, unique features of hiPSC cellular context complicate translation of knowledge from in vivo development and in vitro myogenesis of precommitted progenitors. Several protocols for the directed myogenic differentiation of hiPSCs, including the one that we use in this paper, use gene expression signatures of in vivo somitogenesis as a blueprint for the timing and composition of soluble cues (11–13). In these studies, the undifferentiated tail cone stands in as a proxy for the pluripotent state. However, undifferentiated hiPSCs are most analogous to cells of the primed epiblast, so this comparison may miss key mechanisms of in vitro hiPSC differentiation (3, 5). In addition, hiPSCs have epigenetic priming that can bias subsequent differentiation; this is thought to be due to an interplay of “memory” of the originator somatic cell and culture conditions, including soluble cues and cell density (14–16). The epigenetic priming may be an hiPSC line–dependent contributor to line to line variability.
In this work, we exploit the variation in the myogenic specification of the three hiPSC lines, one robust and two blunted, to decipher differential mechanisms that lead to robust myogenesis. We detail the temporal relationships between the transcriptional, epigenetic, pathway modulation, metabolic, and extracellular factors that lead to robust myogenesis of hiPSCs across three model systems. As a validation of our mechanistic hypotheses, small interfering RNA (siRNA)–mediated perturbation studies suggest that targeted perturbation of a β-catenin cofactor at the onset of differentiation improves initial mesoderm specification, subsequently resulting in better myogenesis.
RESULTS
Robust myogenic specification is hiPSC line dependent
We have carried out myogenic differentiation of three hiPSC lines (TL, SCVI15, and LEPCC3) following a published protocol as described in Materials and Methods and characterized by immunofluorescent (IF) staining for a number of myogenic markers (Fig. 1A) (13). Comparison of gross morphological changes during differentiation showed similar trends—that all the cell lines reached confluence by day 3 and continued to proliferate (fig. S1). IF staining of the cells undergoing differentiation for myogenic markers as a function of time (0 to 30 days) showed cell line–dependent response. Specifically, robust myogenic differentiation was observed in LEPCC3 line (referred to as promyogenic) compared to the SCVI15 and TL lines (referred to as blunted) (Fig. 1A). All cell lines were positive for MYF5 early into differentiation (by day 6). By day 19, the differentiating cells stained positive for desmin, a nonspecific cytoskeletal component found in skeletal, smooth, and cardiac muscle, and the expression level increased with culture time. By day 25, expression of MF20, a skeletal muscle myosin heavy chain, was observed but only in the LEPCC3 line, with increasing expression by day 30. SCVI15 also expressed MF20, but only by day 30; by contrast, the TL line had sparse MF20 expression even after 30 days of differentiation. At days 25 and 30, the LEPCC3 line also showed expression of MYOG and PAX7. These characterizations suggest that LEPCC3 undergoes robust myogenesis compared to the SCVI15 and TL lines.
Fig. 1. Robust myogenic specification is hiPSC line dependent.
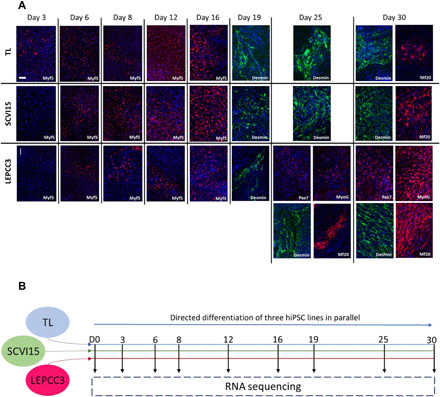
(A) Characterization of myogenic specification of three hiPSC lines with IF staining for multiple markers of myogenesis. Each row is a separate cell line (TL, SCVI15, or LEPCC3), and columns represent days of differentiation (days 3, 6, 8, 12, 16, 19, 25, and 30). Stains are for MYF5 (red), desmin (green), MF20 (red), MYOG (red), PAX7 (red), and nuclei (blue). Each image is labeled with its marker in the lower right corner. SCVI15 and TL cell lines were negative for MYOG and PAX7 at all time points (data not shown). Images are representative of two replicates during parallel culture of all three lines. Scale bar, 100 μm. (B) Schematic of experimental design: Transgene-free myogenic differentiation was induced in three hiPSC lines (TL, SCVI15, and LEPCC3) in parallel according to a published protocol (13) (see also fig. S1). At each time point, myogenic specification of each cell line was characterized by IF staining, and two biological replicates from each line were sent for RNA sequencing.
Gene expression of promyogenic line diverges from the blunted lines
Having observed time- and cell line–dependent myogenic specification with IF characterization, we next carried out longitudinal, whole-transcriptome analyses to better understand the gene regulatory mechanisms that lead to robust myogenic specification in the cellular context of hiPSCs. Specifically, we have carried out longitudinal RNA sequencing of the three lines with temporally differential myogenic specification at nine sequential time points during myogenic differentiation that span the undifferentiated state (day 0) to myogenic specification (day 30) (Fig. 1B).
Quality assessment metrics that demonstrated high-quality raw reads with good alignment to the GRCh38 transcriptome, including complementary DNA (cDNA) and noncoding RNA (ncRNA), are presented in fig. S2. The pipeline for data analysis is detailed in fig. S3. Differential expression analysis identified 16,800 total differentially expressed genes (DEGs) across all cell lines and time points (Fig. 2A and fig. S4). While most of these genes were differentially expressed in multiple cell lines (Fig. 2A), many genes were differentially expressed only in one line; the promyogenic cell line had more unique DEGs compared to the other two lines. While the number of DEGs increased as a function of time in all the cell lines, the promyogenic line had more DEGs at each time point as well as the greatest increase over time in up- and down-regulated coding and noncoding genes (Fig. 2B).
Fig. 2. Gene expression of promyogenic line diverges from the blunted lines.
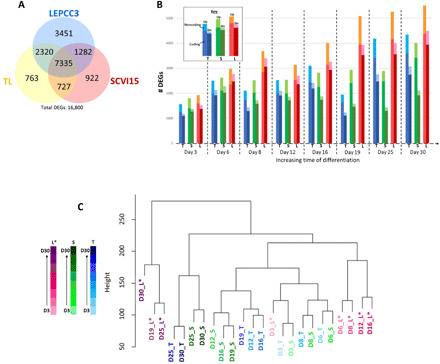
(A) Venn diagram summarizes overlap in number of differentially expressed genes (DEGs) across all time points in the three cell lines (TL, yellow; SCVI15, red; or LEPCC3, blue), with total number of DEGs listed below. (B) Graph shows number of DEGs (y axis) with respect to time (x axis) and cell line (TL, blue; SCVI, green; LEPCC3, red). Each pair of bars represents up-regulated transcripts on the left and down-regulated transcripts on the right. The top-most section of each bar represents the number of noncoding up-regulated (Up) and down-regulated (Dn) transcripts. (C) Hierarchical clustering with respect to DEGs of all time points from all cell lines [LEPCC3 (L*), magenta; SCVI15 (S), green; TL (T), blue]. Darker color represents increasing time of differentiation. Asterisk marks the promyogenic line.
Hierarchical clustering of the samples with respect to DEGs showed the promyogenic line to be increasingly dissimilar from the blunted lines with myogenic differentiation (Fig. 2C), corroborating the IF data (Fig. 1A). At initial time points of differentiation (day 3), all the cell lines clustered together, indicating that they had similar gene expression. However, with increased time of differentiation, the promyogenic samples clustered increasingly farther from the blunted samples, which continued to cluster together. Furthermore, contiguous time points from the same cell line clustered together, indicating consistent differentiation within each line. The cell line–dependent differentiation was further confirmed by R2 correlation coefficients calculated for transcripts per million normalized gene count between each sample. The R2 coefficients that were high between the different cell lines at day 0 progressively decreased with increasing time of differentiation (fig. S5). Together, these analyses showed that the gene expression patterns between the promyogenic and blunted lines began to diverge early in differentiation.
Cell line–dependent gene expression regulates pluripotency and initial lineage commitment
Because the transition from pluripotency to initial lineage specification is a key component of hiPSC differentiation, we next examined the expression of module genes that clustered with key transcriptional regulators of pluripotency (SOX2, PRDM14, POU5F1, KLF4, and NANOG) and initial lineage specification (EOMES, T, HOXA2, HOXB5, and HOXD10) (Fig. 3, A and B). The gene modules were determined by hierarchical clustering with dynamic tree cutting with respect to gene expression across all time points for all three cell lines (17). The functional enrichment of pluripotency and lineage specification of genes supports their roles in regulating pluripotency and differentiation, respectively (fig. S6).
Fig. 3. Cell line–dependent gene expression regulates pluripotency and initial lineage commitment.
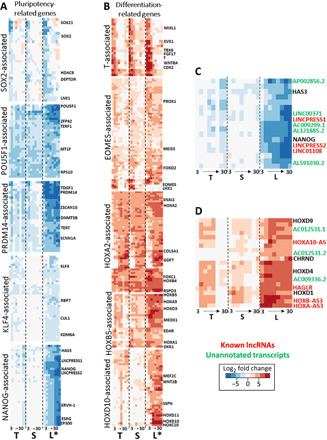
(A) Heatmaps with expression patterns of gene modules associated with regulators of pluripotency: SOX2, POU5F1, PRDM14, KLF4, and NANOG. The modules are determined with dynamic tree cutting of hierarchical clusters with respect to gene expression across all time points and cell lines. Each column of the heatmap represents a cell line, from left to right: T, TL; S, SCVI15; L, LEPCC3 (asterisk marks promyogenic line L*). Within each of the three columns, eight boxes represent time point days 3 to 30. Heatmap colors: Blue represents down-regulated and red represents up-regulated L2FC gene expression with respect to each cell line’s respective day 0 undifferentiated value. (B) Heatmaps with expression patterns of gene modules associated with key regulators of differentiation: T and EOMES are crucial regulators of mesendoderm specification, while HOXA2, HOXB5, and HOXD10 are representative of anterior to posterior HOX family genes. (C) Heatmap detailing the noncoding RNA (ncRNA) transcripts that cluster near NANOG: red, known lncRNAs; green, unannotated transcripts. (D) Heatmap detailing the noncoding ncRNA transcripts that cluster near HOXD9: red, known lncRNAs; green, unannotated transcripts.
The modules of genes associated with SOX2, POU5F1, and PRDM14 were down-regulated across all the cell lines, albeit with subtle differences (Fig. 3A). Other regulators of pluripotency in these modules include SOX21, DEPTOR, ZFP42 (REX1), TDGF1, and ZSCAN10. The telomere regulators TERF1 and TERT within the modules were also found to be down-regulated in all three cell lines. By contrast, the KLF4- and NANOG-associated modules were down-regulated in the promyogenic line but less so in the other cell lines. The NANOG-associated module includes long noncoding RNAs (lncRNAs) that play a role in the regulation of pluripotency, such as LINCPRESS1/2 and LINC01108 (embryonic stem cell-associated 1) (18, 19). Together, these expression patterns indicate cell line–dependent transition from pluripotency to lineage restriction, particularly with respect to genes that may play a role in NANOG-mediated regulation.
The modules of genes associated with key regulators of initial differentiation also showed time- and cell line–dependent expression (Fig. 3B). The T-associated module of genes includes regulators of mesendoderm specification including MSGN1, FGF17, CDX1/2, and WNT8A. While this module was up-regulated early in differentiation in all three cell lines, these genes were up-regulated for a longer duration in the promyogenic line compared to the blunted lines. The EOMES-associated genes were generally up-regulated in all three cell lines; however, the genes that clustered most closely with EOMES, including LHX1, a transcription factor regulating endoderm specification, were up-regulated only in the blunted lines but not in the promyogenic line.
We also analyzed the HOX genes, HOXA2 (anterior), HOXB5 (middle), and HOXD10 (posterior), as the HOX family genes play a role in anterior-to-posterior axis determination in embryonic development (20). All the cell lines showed up-regulation of genes in the anterior HOX2A-associated module and the middle HOXB5-associated module. These modules include the genes SNAI2, an epithelial-mesenchymal transition regulator, and MEOX1. In contrast, the posterior HOXD10-associated module was up-regulated in the promyogenic line compared to the blunted lines. This module also includes multiple regulatory lncRNAs, including HAGLR and HOXA10-AS (21). We also noticed the presence of multiple unannotated transcripts with synchronous expression patterns, several of which are putative lncRNAs (Fig. 3, C and D).
Gene expression of chromatin-modifying complex components is cell line dependent
Transcription factor enrichment analysis using the pluripotency- and differentiation-associated gene modules (Fig. 3) indicated the involvement of polycomb repressor complexes 1 and 2 (PRC1/2) in gene regulation, as multiple PRC1/2 components were significantly enriched (fig. S7) (22). PRC1 and PRC2 are active in histone modification including epigenetic priming in pluripotent cells (23, 24). Several PRC1 targeting components have cell line–dependent expression, including KDM2B, which was found to be down-regulated more in the promyogenic line compared to the blunted lines. The enzymatic components of PRC1 also have cell line–dependent expression, with RING1 preferentially expressed in the promyogenic line compared to RNF2 (Fig. 4A). Although the core components of PRC2 were significantly enriched upstream of both pluripotency- and lineage specification–related genes (fig. S7), they were not differentially expressed in any of the cell lines (Fig. 4B). However, targeting component JARID2, involved in pluripotency maintenance, was down-regulated in the promyogenic, but not in blunted, lines (25).
Fig. 4. Gene expression of chromatin-modifying complex components is cell line dependent.
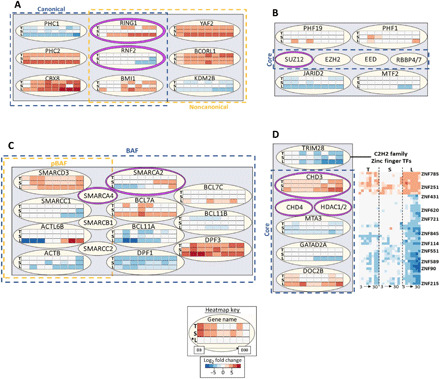
(A) Selected components of polycomb repressor complex 1 (PRC1). Components of the canonical complex are inside the dotted blue box, and components of the noncanonical complex are inside the dotted yellow box. The enzymatic components are ringed in purple. Each row in every three-row heatmap represents one cell line, from top to bottom: T, TL; S, SCVI15; L, LEPCC3 (asterisk marks promyogenic line L*). Each row has eight boxes representing time points days 3 to 30. Heatmap colors: Blue represents down-regulated and red represents up-regulated L2FC gene expression with respect to each cell line’s respective day 0 undifferentiated value. (B) Selected components of PRC2. Core components are inside the dotted blue box, and enzymatic component is ringed in purple. (C) Selected components common to pBAF and BAF complexes. All the components shown are part of BAF (dotted blue box), while some are also in pBAF (dotted yellow box). Enzymatic components are ringed in purple. (D) Selected components of the nucleosome remodeling and deacetylase (NuRD) complex, with core components in the dotted blue box and enzymatic components ringed in purple. TRIM28 is shown with several associated zinc finger transcription factors (TFs).
BRG1/BRM associated factor (BAF)/polybromo associated BAF (pBAF) and NuRD (nucleosome remodeling and deacetylase) are chromatin-modifying complexes that operate largely at the nucleosome level (26, 27). Several components of the BAF family complexes showed a cell line–dependent expression, including adenosine 5′-triphosphate (ATP)–dependent enzyme SMARCA2 and targeting components BCL7A and BCL11A (Fig. 4C). NuRD complex targeting component TRIM28, a regulator of pluripotency, was more down-regulated in the promyogenic than in blunted lines. In addition, multiple TRIM28-associated known and unannotated zinc finger transcription factors have also exhibited cell line–dependent expression (Fig. 4D) (28). In short, we observed cell line–dependent expression of ancillary components that mediate targeting, rather than cell line–dependent expression of core or enzymatic components, with a few exceptions.
Temporally differential expression of mesendoderm regulators precedes myogenic specification
On the basis of our observation of time- and cell line–dependent expression of key genes that regulate pluripotency (NANOG, POU5F1, KLF4, and SOX2) and initial lineage specification (EOMES and T) (Fig. 3), we next tracked the downstream lineage specification in the promyogenic and blunted lines (Fig. 5A). The blunted lines showed an initial specification to endoderm-like fate (LHX1 and SOX17) and cardiogenic mesoderm (GATA4 and NKX2-5). Concomitant with this early commitment, the blunted lines expressed genes associated with endoderm-derived liver (SERPINA1 and HNF4A) and cardiac morphogenesis (HAND1) at later time points of differentiation. In contrast, the promyogenic line showed an increased expression of genes associated with somitogenesis (MEOX1), intermediate mesoderm (OSR1), and paraxial mesoderm (PAX3 and PDGFRA), with downstream expression of the corresponding derivatives of urogenital tract development (WT1) and terminal myogenesis (MYOG).
Fig. 5. Time- and hiPSC line–dependent gene expression leads to robust myogenic specification.
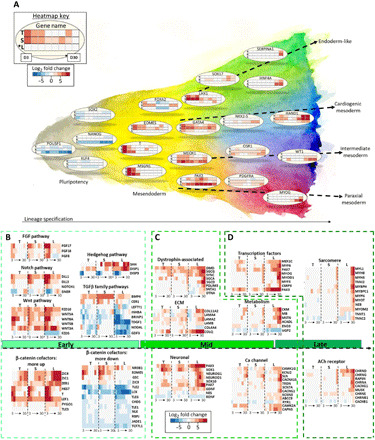
(A) Schematic of cell line–dependent lineage specification. From left to right: Key regulators of pluripotency, mesendoderm specification, and finally key genes that represent mesendoderm derivatives. Each row in every three-row heatmap represents one cell line, from top to bottom: T, TL; S, SCVI15; L, LEPCC3 (asterisk marks promyogenic line L*). Each row has eight boxes representing time points days 3 to 30. Heatmap colors: Blue represents down-regulated and red represents up-regulated L2FC gene expression with respect to each cell line’s respective day 0 undifferentiated value. (B) At the outset of differentiation, preferential up-regulation of key FGF, Notch, Wnt, and down-regulation of TGFβ family pathway components in the promyogenic line, with synchronous cell line–dependent up- and down-regulation of multiple transcriptional cofactors of β-catenin. (C) In mid-differentiation, preferential expression in the promyogenic line of groups of genes known to promote myogenic differentiation and function (dystrophin-associated, ECM components, and neuronal genes precede myogenic transcription factors, metabolic factors, and calcium handling). (D) Late in differentiation, there is up-regulation of genes related to myogenesis (sarcomeric and AChR components), predominantly in the promyogenic line.
Time- and cell line–dependent expression of pathway components and β-catenin cofactors
We next examined the gene expression of members of major signaling pathways (Fig. 5B). The genes in each pathway or functional group were curated through a combination of HUGO gene nomenclature committee (HGNC) families and literature search (29). Consistent with exposure to CHIR during early differentiation, we observed up-regulation of members of the Wnt pathway (including WNT3A). We also observed up-regulation of Notch, Hedgehog, and FGF8, FGF17, and FGF18 pathway members in all cell lines. However, this up-regulation was for a longer duration in the promyogenic compared to the blunted lines. In contrast, although all cell lines had identical exposure to the exogenous small molecule LDN, transforming growth factor–β (TGFβ) family members were more down-regulated in the promyogenic line and up-regulated in the blunted lines. CER1 and NODAL, secreted ligands of the TGFβ superfamily that have been shown to promote endoderm differentiation, were more up-regulated early in differentiation in the lines with blunted myogenesis (30).
Despite the exposure to same exogenous factors (e.g., small molecules), the differentiation and pathway modulation was cell line dependent, suggesting the involvement of other factors such as epigenetic state and availability of β-catenin cofactors, which could contribute to initial lineage specification (24, 31–33). To this end, we have assessed the expression profile of the β-catenin cofactors and β-catenin targets. Genes that were more up-regulated in the promyogenic line include transcription factors PYGO, ZIC1/4, and LEF1, Notch-regulated transcription factor HES7, and T. Genes that were more down-regulated in the promyogenic line include TLE1, TLE2, ZIC3, and TLE6, as well as EOMES and GSC (Fig. 5B).
Cell line–dependent expression of extrinsic and intrinsic factors precedes robust myogenesis
By middle to late differentiation time points, the promyogenic line has preferential expression of several intra- and extracellular components involved in myogenic specification (Fig. 5C). Genes related to extracellular matrix (ECM) molecules relevant to skeletal muscle, such as LAMA2 and COLQ [a structural component of the acetylcholine receptor (AChR)] as well as components of the dystrophin complex, which anchors myocytes to the ECM, are more up-regulated in the promyogenic lines. Muscle-specific metabolic factors, including creatinine kinase (CKM) (a clinically used marker of muscle injury), myoglobin (involved in oxygen metabolism), and myostatin (involved in skeletal muscle cell signaling), were also more up-regulated in the promyogenic line. Genes related to calcium handling, including RYR1, a component of a sarcoplasmic reticulum calcium channel that plays a role in muscle contraction, were likewise more up-regulated in the promyogenic line. Several genes related to neural development, such as NEUROG1, NEUROD1, and NES, were also found to be preferentially up-regulated in the promyogenic cell line (Fig. 5C).
Concomitantly, by late differentiation time points, genes involved in downstream myogenic maturation were more up-regulated in the promyogenic than the blunted lines (Fig. 5D). Transcription factors that play a role in paraxial mesoderm development (e.g., PAX3 and PAX7) and myogenic regulatory factors (e.g., MYOD1 and MYOG) were more up-regulated in the promyogenic line. Transcription factor expression preceded up-regulation of skeletal muscle-specific sarcomeric heavy- and light-chain myosins and Z-disc components. Components of the fetal neuromuscular junction AChR, such as CHRND and CHRNG, were up-regulated in the promyogenic line by early differentiation time points, and components of the more mature AChR, such as CHRNA, CHRNB1, and MUSK, were up-regulated by late time points.
Transcriptional regulation of initial lineage commitment is time and cell line dependent
We further delineated the temporal relationships between processes that correlated with robust myogenic specification in the promyogenic line using the Short Time-series Expression Miner (STEM) (34). Seven statistically significant temporal profiles and their corresponding Gene Ontology categories are shown in Fig. 6A. ECM-related categories steadily increased (profile 39) over the course of myogenic specification, while categories related to RNA and DNA processing steadily decreased (profile 10). Glycolysis (profile 21) decreased over the course of differentiation, consistent with loss of pluripotency (35), and increased only in late differentiation time points, synchronous with the appearance of myoblasts. Profile 37, enriched for cilia-related processes, peaked in the latter part of differentiation, consistent with the role of primary cilia in early myoblast commitment (36). Profiles related to somitogenesis (profile 47) peaked early in differentiation, while those related to embryonic skeletal muscle development (profile 38) increased gradually over early and mid-differentiation. Last, genes related to contractile myofibers (profile 24), indicating more mature skeletal muscle, increased sharply only by late differentiation time points.
Fig. 6. Transcriptional regulation of initial lineage commitment is time and cell line dependent.
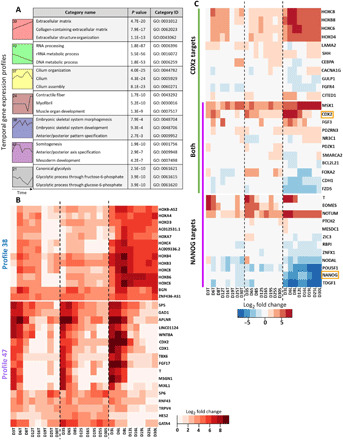
(A) Seven significant temporal profiles (profiles: 39, red; 10, green; 37, yellow; 24, brown; 38, blue; 47, purple; 21, gray) as well as select corresponding enriched Gene Ontology categories were determined with STEM software. (B) Comparison of gene expression for sets of genes driving enrichment of categories in profile 38 (top heatmap) and profile 47 (bottom heatmap) showed cell line–dependent expression, including multiple key transcription factors. (C) Transcription factors CDX2 and NANOG had cell line–dependent expression (orange boxes), as did transcriptional targets of NANOG (purple), CDX2 (green), and both (overlap purple and green). TF targets were determined using the ChEA database in Enrichr (52).
We next compared gene expression in all three cell lines for temporal profiles that increased markedly in early differentiation (Fig. 6B), because mesoderm commitment preceded downstream myogenesis in the promyogenic line (Fig. 5A). Enrichment of terms related to embryonic skeletal development (profile 38) was driven largely by expression of anterior to middle HOX family transcription factors. The HOX genes have more robust and earlier up-regulation in the promyogenic line compared to the blunted lines (by day 3 versus day 6). Profile 47 is enriched for terms related to somitogenesis and mesoderm development. Key transcription factors in this temporal profile, such as T, TBX6, MSGN1, and CDX1/2, are increased in early differentiation in all three cell lines but have more robust and longer duration of up-regulation in the promyogenic line.
As CDX2 was identified in profile 47, and because NANOG and CDX2 have been shown to jointly regulate differentiation to different mesoderm subtypes at the exit from pluripotency (37), we compared the expression of transcriptional targets of NANOG and CDX2 (Fig. 6C). We observed cell line–dependent expression of NANOG and CDX2 (orange boxes) as well as multiple transcriptional targets at the outset of differentiation. These targets include several genes that are important in lineage specification, including T, EOMES, and NODAL (regulated by NANOG); FGF3, CDH1, and FOXA2 (regulated by both); SHH; and multiple middle HOX genes (regulated by CDX2). Together, this indicates time-dependent transcription factor signatures that distinguish the promyogenic line from the blunted lines in early differentiation time points.
β-Catenin cofactors integrate exogenous cues and pathway, epigenetic, and transcriptional mechanisms
On the basis of the observations and data analyses described above, we posit that the cell line–dependent expression of β-catenin cofactors (Fig. 5B) affects initial lineage specification by integrating exogenous cues (fig. S1) and transcriptional (Fig. 3, A and B), epigenetic (Figs. 3, C and D, and 4), and signaling pathways (Fig. 5B) (16, 31, 33, 38–40). In the promyogenic line, there is a longer duration of T expression and no EOMES expression compared to the blunted lines, resulting in initial specification to the mesoderm lineage, which, in turn, leads to robust downstream myogenesis (Fig. 5A). We therefore focused on initial lineage specification to mesoderm versus endoderm based on expression of T and EOMES, both of which are transcriptional targets of β-catenin. The network diagram summarizes ways in which exogenous cues, signaling pathways, epigenetics, and pluripotency transcription factors interact, through β-catenin cofactors, to potentially modulate β-catenin transcriptional activity (including transcription of T and EOMES) (Fig. 7A). The key interactions are detailed in fig. S8, with protein-protein interaction data obtained from STRINGdb (41).
Fig. 7. Initial ZIC3 knockdown leads to enhanced terminal myogenic specification.
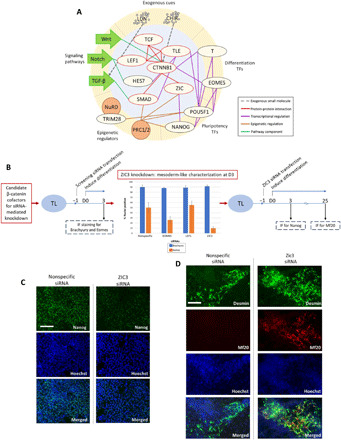
(A) In our network of initial lineage specification, transcriptional cofactors of β-catenin mediate cross-talk between signaling pathway activity, epigenetics, transcription factors, and exogenous cues. CTNNB1 (β-catenin) and cofactors that physically interact are highlighted in red. Gray dotted line, exogenous small-molecule CHIR and LDN (represented with gray circles) that modulate Wnt and TGF-β signaling pathways, respectively; red, protein-protein interaction (experimentally determined from STRINGdb); green, signaling pathways and members; orange, epigenetic regulator targets; purple, transcriptional regulation (see also fig. S8). (B) Schematic of experimental design for siRNA-mediated knockdown (KD) screen (left) and ZIC3 KD (left) to enhance myogenic specification in hiPSC line with blunted myogenesis (TL). (C) Representative images showing attenuated NANOG expression at day 3 of differentiation in ZIC3 siRNA-mediated KD condition compared to nonspecific siRNA control. IF staining for NANOG (green) and nuclei (blue). Scale bar, 100 μm. (D) Representative images showing enhanced myogenic specification at day 25 of differentiation in hiPSC line with blunted myogenesis (TL) due to siRNA KD of ZIC3 at the outset of differentiation compared to nonspecific siRNA condition. IF staining for desmin (green), MF20 (red), and nuclei (blue). Scale bar, 100 μm.
ZIC3 knockdown leads to enhanced downstream myogenic specification
Guided by the network model (Fig. 7A), we hypothesized that targeted perturbation of a β-catenin cofactor at the outset of differentiation could improve initial mesoderm specification, leading to enhanced downstream myogenic specification. To test this, we carried out an siRNA-mediated knockdown screen of multiple candidate β-catenin cofactors (fig. S9) in a blunted cell line (TL) (Fig. 7B). On the basis of an siRNA screen, knockdown of the gene ZIC3 showed less endoderm-like differentiation while exhibiting high T and attenuated EOMES expression at 3 days after differentiation (fig. S10) (40, 42, 43). Hence, we have evaluated the effect of ZIC3 knockdown at the outset of differentiation on myogenic specification. In accordance with the siRNA treatment, we observed decreased NANOG expression by IF staining by day 3 of differentiation compared to the scrambled siRNA control (Fig. 7C) (43). By 25 days of differentiation, we observed enhanced MF20 expression by IF staining in the ZIC3 knockdown condition compared to nonspecific siRNA control, suggesting improved myogenic specification (Fig. 7D).
DISCUSSION
In this study, we compared hiPSCs with line-to-line variation in differentiation to better understand the temporal mechanisms of myogenic specification of hiPSCs. To this end, we have generated a unique transcriptomic dataset that captures a longitudinal head-to-head comparison of multiple hiPSC lines with differential myogenic commitment. We detailed gene expression patterns related to transcriptional, signaling pathway, and epigenetic regulation that lead to robust myogenic specification. At the outset of differentiation, we observed a divergence in gene expression between the cell lines that suggests differences in initial germ layer specification. We hypothesized that transcriptional cofactors of β-catenin play a role in integrating multiple mechanisms to affect lineage specification and showed that perturbation of one such cofactor, ZIC3, led to improved myogenic specification in a blunted line.
Despite the fundamental differences between in vitro differentiation of hiPSCs and in vivo embryonic development, a comparison can be helpful to understand our observations, as well as underscore the importance of our study within the hiPSC context. We consider initial induction of differentiation in vitro with the analogy of in vivo primitive streak (PS) formation, because undifferentiated hiPSCs are thought to be most analogous to the primed epiblast (3, 5, 12). During embryogenesis, the epiblast gives rise to the PS, which is pan-positive for T; from the PS, germ layers arise in a time- and space-dependent fashion, with endoderm arising first (early anterior PS) followed by mesoderm (late posterior PS) (44). Likewise, on induction of hiPSC differentiation in vitro, we observe that all cell lines have up-regulation of T; however, T expression is prolonged only in the promyogenic line, which could be attributed to these cells committing to a late PS-like fate compared to an early streak fate in the blunted lines. Early PS derivatives include endoderm, cardiogenic, and head mesoderm, while intermediate and paraxial mesoderm are derived from the late PS (45), and these stages were captured by the differentiating hiPSCs in a time-dependent manner. The cell line–dependent pattern of HOX gene expression (posterior HOX genes up-regulated preferentially in the promyogenic line) and signaling pathway modulation (increased TGFβ component expression in the blunted lines) also concurs with the anterior versus posterior PS commitment analogy. The lines with blunted myogenesis had persistent NANOG expression, which fits with the smaller total number of DEGs in these lines compared to the promyogenic line and may indicate an earlier fate even by late time points of differentiation (37).
Although analogy with early embryonic development can frame certain observations, there are aspects unique to hiPSCs such as that the differentiation is heterogeneous in terms of cell subpopulations that arise. We observe a total of 16,800 DEGs in all time points from all the cell lines. This represents differential expression of about 30% of the possible transcripts and likely reflects heterogeneity, as initial specification gives rise to multiple derivatives with increasing time of differentiation. The promyogenic line has more up- and down-regulated genes at all time points as well as more unique genes compared to the blunted lines, suggesting the presence of different subpopulations. Neural “contaminants” have been reported in several protocols for the myogenic differentiation of hPSCs, including the protocol that we use (13). Nevertheless, it is plausible that these subpopulations play a role in promoting myogenic specification; in vitro studies have shown enhanced myogenesis of primary myoblasts with motor neuron coculture (46). Similarly, the appearance of Pax7+ satellite cells in conjunction with MyoG+ terminally specified myogenic cells represents a heterogeneous cell population that arises within a niche. The heterogeneous differentiation of hiPSCs makes it difficult to disambiguate specific details of distinct cell subpopulations. Although cell sorting or single-cell sequencing may disrupt cell-cell and cell-ECM interactions that can influence lineage specification, future studies should consider these modalities to better understand subpopulation heterogeneity.
Our data showed high correlation among the biological replicates from the same cell line, even with increasing time of differentiation, demonstrating self-consistency during parallel differentiation. Therefore, trends in gene expression, especially synchronous expression of groups of genes as revealed by our longitudinal approach, provide insights into lineage specification. For example, cell line–dependent temporal variation in transcription factor and signaling pathway component expression shows differential initial lineage specification between the cell lines. Furthermore, our longitudinal data enable inference of causality. In addition, unbiased analysis techniques to identify modules of genes with similar expression patterns can help contextualize potential function of unannotated transcripts, as in the case of unannotated lncRNAs that cluster with NANOG and the HOX genes as well as multiple ZNF transcription factors that associate with TRIM28. Future experiments could explore the roles of these unannotated genes, especially as they may relate to epigenetic regulation. Last, our analyses and perturbation experiment suggest the key role played by the β-catenin cofactors at the outset of differentiation and on downstream targets for myogenic differentiation. Together, our study is a framework to better understand and potentially improve myogenic specification of hiPSCs.
MATERIALS AND METHODS
Pluripotent hiPSC culture
hiPSC lines (LEPCC3, SCVI15, and TL) were derived from peripheral blood mononuclear cells from healthy individuals. The SCVI15 and TL lines were obtained through material transfer agreement from the Stanford University and Ann & Robert H. Lurie Children’s Hospital Chicago. The LEPCC3 line was generated jointly with the Salk Institute. Pluripotent hiPSC colonies were plated onto six-well plates that had been coated with Matrigel (Corning) according to the manufacturer’s instructions overnight at 4°C. Maintenance medium mTeSR1 (STEMCELL Technologies) was refreshed daily. hiPSCs were passaged at ~70% confluence using ReLeSR (STEMCELL Technologies) according to the manufacturer’s instructions to maintain pluripotent colonies. From frozen vials, pluripotent colonies were passaged twice before seeding for differentiation. All three cell lines were cultured in parallel.
Myogenic differentiation
Pluripotent hiPSC colonies were seeded for differentiation by adding 500 μl of Versene (0.5 mM EDTA solution, Gibco) per well in a six-well plate and incubating at 37°C for 9 min to detach cells as single cells. These were then replated as single cells on Matrigel-coated plates as above. Undifferentiated hiPSCs were plated at a seeding density of 20,000 cells/cm2 in mTeSR with 1% penicillin-streptomycin (Gibco) and 2 μM Thiazovivin (Selleck Chemicals) for 24 hours to allow attachment. Differentiation was induced 24 hours after plating following the protocol by Chal et al. (11). Briefly, five differentiation medium conditions were used over the course of 30 days of myogenic differentiation: days 1 to 3: Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco) with 1% NEAA (non-essential amino acid) (Gibco), 1% GlutaMAX (Gibco), 1% penicillin-streptomycin (Gibco), 1% ITS (insulin-transferrin-selenium) supplement (Gibco), CHIR99021 (3 μM) (Selleck Chemicals), and LDN-193189 (0.5 μM) (Miltenyi Biotec); days 4 to 6: DMEM/F12 with 1% NEAA, 1% GlutaMAX, 1% penicillin-streptomycin, 1% ITS supplement, CHIR99021 (3 μM), LDN-193189 (0.5 μM), and fibroblast growth factor (FGF) (20 ng/ml) (R&D Systems); days 7 and 8: DMEM/F12, 15% KOSR (KnockOut Serum Replacement) (v/v) (Gibco), 1% NEAA, 1% GlutaMAX, 1% penicillin-streptomycin, HGF (hepatocyte growth factor) (10 ng/ml) (R&D Systems), IGF (insulin-like growth factor) (2 ng/ml) (R&D Systems), FGF (fibroblast growth factor) (20 ng/ml), LDN (0.5 μM), and β-mercaptoethanol (0.1 mM); days 9 to 12: DMEM/F12, 15% KOSR, 1% NEAA, 1% GlutaMAX, 1% penicillin-streptomycin, IGF (2 ng/ml), and β-mercaptoethanol (0.1 mM); days 13 to 30: DMEM/F12, 15% KOSR, 1% NEAA, 1% GlutaMAX, 1% penicillin-streptomycin, HGF (10 ng/ml), IGF (2 ng/ml), and β-mercaptoethanol (0.1 mM). Medium was changed daily for the first 12 days, followed by half medium change every day for the remainder of the differentiation to day 30. All three hiPSC lines were cultured in parallel for transcriptomic studies as well as corresponding IF staining characterization.
IF staining
IF staining was performed using the following primary antibodies: PAX7 (1∶100; Developmental Studies Hybridoma Bank), MF20 (1∶200; Developmental Studies Hybridoma Bank), MYF5 (1∶200; Santa Cruz Biotechnology), desmin (1∶200; Abcam), MYOG (1∶100; Developmental Studies Hybridoma Bank), Brachyury (1:100; R&D Systems, AF-2085), EOMES (1:100; R&D Systems, MAB6166), and NANOG (1:200; Santa Cruz Biotechnology). The following secondary antibodies were used: goat anti-rat Alexa 546 (1∶200; Life Technologies), goat anti-mouse Alexa 488 (1∶250; Life Technologies), and goat anti-rabbit Alexa 546 (1∶200; Life Technologies). For IF staining of cells grown on tissue culture plates, cells were fixed in 4% paraformaldehyde for 10 min at room temperature. Immediately before staining, the cells were permeabilized with 0.1% (v/v) Triton X-100 and blocked with 3% (w/v) bovine serum albumin (BSA) for 30 min. Cells were stained with primary antibodies in dilution ratios as listed above in 1% BSA overnight at 4°C, washed three times with phosphate-buffered saline, and incubated with secondary antibodies for 1 hour at room temperature. The nuclei were stained with Hoechst 33342 (2 μg/ml; Life Technologies) for 5 min at room temperature. Imaging was performed using a fluorescence microscope (Carl Zeiss; Axio Observer A1).
RNA extraction, quality assessment, library preparation, and sequencing
RNA samples were collected for nine time points with two biological replicates for each of three cell lines using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s instructions. At the University of California San Diego (UCSD) Institute for Genomic Medicine, RNA integrity number (RIN) scores for each sample were calculated using Agilent 4200 TapeStation instrument and samples with RIN lower than 8 were excluded from downstream sequencing. Library preparation was carried out with an Illumina TruSeq Stranded mRNA kit for poly-A selection. Samples were sequenced on Illumina HiSeq 4000. One hundred–base pair paired-end reads with about 30 million reads per sample were generated.
RNA sequencing data quality check, read alignment, and differential expression analysis
The quality of raw reads was assessed using FastQC (Babraham Bioinformatics), and quality metrics for multiple samples were compiled with MultiQC (47). Alignment to the human transcriptome (Ensembl version GRCh38.p10) that consisted of cDNA and ncRNA transcripts was performed using Salmon (48). Summation of transcript read counts to gene counts was done using tximport (49). Differential gene expression analysis was carried out using DESeq2 (50), with cutoffs of Padj ≤ 0.005 and L2FC (log2 fold change) ≥ 1. Downstream analyses were performed using the R statistical computing software (51).
siRNA screen
Wells of a 384-well plate were coated with Matrigel (Corning) per the manufacturer’s instructions at 4°C overnight. At time of cell seeding, Matrigel was aspirated, 5 μl of 1% Lipofectamine RNAiMAX transfection reagent (Invitrogen) in Opti-MEM (Gibco) was added per well, and 2.5 μl of 0.5 μM siRNA was added per well, with spin down of plate to ensure settling of reagents. siRNAs were purchased from Dharmacon Inc. as pool of four siRNA targeting sequences per gene. hiPSCs (TL cell line) were seeded as single cells at a seeding density of 20,000 cells/cm2, as above, for myogenic differentiation. Final volume per well was 50 μl, including cells, for final siRNA concentration of 25 nM. Twenty-four hours were allowed for cell attachment and transfection before removing plating medium and inducing myogenic differentiation, as above. At day 3, cells were fixed and permeabilized for immunostaining as above and stained for Brachyury (1:100; R&D Systems, AF-2085) and EOMES (1:100; R&D Systems, MAB6166), with secondary and nuclear staining, as above. Fluorescence was imaged with an automated microscope (ImageXpress, Molecular Devices) and quantified with the MetaXpress software (Molecular Devices). There were two biological replicates per experimental condition and four images per well for a total of eight quantified images per experimental condition for average and SD calculations.
RNA sequencing data
Raw and processed data are deposited to the Gene Expression Omnibus repository with accession number GSE161025.
Statistical analysis
Alignment of raw reads to the transcriptome was performed using a quasi-mapping and dual-phase inference method as implemented by Salmon, gene-level estimation of transcript abundance was performed with tximport, and differential expression analysis was performed using a hypergeometric distribution as implemented by DESeq2 with cutoffs of Padj ≤ 0.005 and L2FC ≥ 1 (see above). IF staining image quantification is presented as means ± SD. STEM analysis software calculates gene set enrichment analysis using a hypergeometric distribution, as described in (34).
Supplementary Material
Acknowledgments
We thank A. Caldwell of the Subramaniam laboratory and Y. Shih of the Varghese laboratory. We also thank S. Spiering, M. Wu, and M. Missinato of the Colas laboratory for help with siRNA experiments. We thank UCSD Institute for Genomic Medicine for help with RNA sequencing. Funding: We acknowledge research funding support from the California Institute of Regenerative Medicine (RT3-07907 to S.V.), the NIH (R01 LM012595, U01 CA198941, U01 DK097430, R01 DK109365, R01 HD084633, and R01 HL108735 to S.S.), the NSF (STC CCF-0939370 to S.S.), the Joan and Irwin Jacobs Endowment (to S.S.), P01HL141084 (to M.M.), and the Joan and Sanford I. Weill Scholars Endowment (to M.M.). Author contributions: P.N., S.V., and S.S. designed all experiments and analyses. P.N. carried out all experiments and bioinformatics analysis. M.M. and A.C. assisted with validation experiments. S.S. and S.V. supervised the project. P.N. wrote the first draft of the manuscript with help from S.V. and S.S. All authors have reviewed and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/12/eabf7412/DC1
REFERENCES AND NOTES
- 1.Wu S. M., Hochedlinger K., Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 13, 497–505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiwlawat N., Lynch E., Jeffrey J., Van Dyke J. M., Suzuki M., Current progress and challenges for skeletal muscle differentiation from human pluripotent stem cells using transgene-free approaches. Stem Cells Int. 2018, 6241681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chal J., Pourquié O., Making muscle: Skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Trounson A., DeWitt N. D., Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194–200 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Bilic J., Belmonte J. C. I., Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells 30, 33–41 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L., Webster S. G., Miller S. C., Webster C., Plasticity of the differentiated state. Science 230, 758–766 (1985). [DOI] [PubMed] [Google Scholar]
- 7.Hwang Y., Phadke A., Varghese S., Engineered microenvironments for self-renewal and musculoskeletal differentiation of stem cells. Regen. Med. 6, 505–524 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Mukund K., Subramaniam S., Skeletal muscle: A review of molecular structure and function, in health and disease. WIREs Syst. Biol. Med. 12, e1462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young C. S., Hicks M. R., Ermolova N. V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J. A., Kohn D. B., Nakano A., Nelson S. F., Miceli M. C., Spencer M. J., Pyle A. D., A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 18, 533–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang Y., Suk S., Lin S., Tierney M., Du B., Seo T., Mitchell A., Sacco A., Varghese S., Directed in vitro myogenesis of human embryonic stem cells and their in vivo engraftment. PLOS ONE 8, e72023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P., Tassy O., Vincent S., Miyanari A., Bera A., Garnier J.-M., Guevara G., Hestin M., Kennedy L., Hayashi S., Drayton B., Cherrier T., Gayraud-Morel B., Gussoni E., Relaix F., Tajbakhsh S., Pourquié O., Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 33, 962–969 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Xi H., Fujiwara W., Gonzalez K., Jan M., Liebscher S., Handel B. V., Schenke-Layland K., Pyle A. D., In vivo human somitogenesis guides somite development from hPSCs. Cell Rep. 18, 1573–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chal J., Al Tanoury Z., Hestin M., Gobert B., Aivio S., Hick A., Cherrier T., Nesmith A. P., Parker K. K., Pourquié O., Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nat. Protoc. 11, 1833–1850 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Boland M. J., Nazor K. L., Loring J. F., Epigenetic regulation of pluripotency and differentiation. Circ. Res. 115, 311–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W., Plath K., Smith A., Epigenetic resetting of human pluripotency. Development 144, 2748–2763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang G., Zhang Y., Genetic and epigenetic variations in iPSCs: Potential causes and implications for application. Cell Stem Cell 13, 149–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P., Zhang B., Horvath S., Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Fico A., Fiorenzano A., Pascale E., Patriarca E. J., Minchiotti G., Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cell. Mol. Life Sci. 76, 1459–1471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keshavarz M., Asadi M. H., Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J. 286, 2611–2623 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Pineault K. M., Wellik D. M., Hox genes and limb musculoskeletal development. Curr. Osteoporos. Rep. 12, 420–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar B. D., Krumlauf R., HOXs and lincRNAs: Two sides of the same coin. Sci. Adv. 2, e1501402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aloia L., Stefano B. D., Croce L. D., Polycomb complexes in stem cells and embryonic development. Development 140, 2525–2534 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Illingworth R. S., Hölzenspies J. J., Roske F. V., Bickmore W. A., Brickman J. M., Polycomb enables primitive endoderm lineage priming in embryonic stem cells. eLife 5, e14926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Jones A., Sun C.-W., Li C., Chang C.-W., Joo H.-Y., Dai Q., Mysliwiec M. R., Wu L.-C., Guo Y., Yang W., Liu K., Pawlik K. M., Erdjument-Bromage H., Tempst P., Lee Y., Min J., Townes T. M., Wang H., PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprograming. Stem Cells 29, 229–240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho P. J., Lloyd S. M., Bao X., Unwinding chromatin at the right places: How BAF is targeted to specific genomic locations during development. Development 146, dev178780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hota S. K., Bruneau B. G., ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882–2897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oleksiewicz U., Gładych M., Raman A. T., Heyn H., Mereu E., Chlebanowska P., Andrzejewska A., Sozańska B., Samant N., Fąk K., Auguścik P., Kosiński M., Wróblewska J. P., Tomczak K., Kulcenty K., Płoski R., Biecek P., Esteller M., Shah P. K., Rai K., Wiznerowicz M., TRIM28 and Interacting KRAB-ZNFs control self-renewal of human pluripotent stem cells through epigenetic repression of Pro-differentiation genes. Stem Cell Rep. 9, 2065–2080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braschi B., Denny P., Gray K., Jones T., Seal R., Tweedie S., Yates B., Bruford E., Genenames.org: The HGNC and VGNC resources in 2019. Nucleic Acids Res. 47, D786–D792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempf H., Olmer R., Haase A., Franke A., Bolesani E., Schwanke K., Robles-Diaz D., Coffee M., Göhring G., Dräger G., Pötz O., Joos T., Martinez-Hackert E., Haverich A., Buettner F. F. R., Martin U., Zweigerdt R., Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun. 7, 13602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony C. C., Robbins D. J., Ahmed Y., Lee E., Nuclear regulation of Wnt/β-catenin signaling: It’s a complex situation. Genes 11, 11080886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galceran J., Sustmann C., Hsu S.-C., Folberth S., Grosschedl R., LEF1-mediated regulation of Delta-like1 links Wnt and Notch signaling in somitogenesis. Genes Dev. 18, 2718–2723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu F.-I., Sun Y.-H., Wei C.-Y., Thisse C., Thisse B., Tissue-specific derepression of TCF/LEF controls the activity of the Wnt/β-catenin pathway. Nat. Commun. 5, 5368 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Ernst J., Bar-Joseph Z., STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Nuebel E., Daley G. Q., Koehler C. M., Teitell M. A., Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11, 589–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu W., Asp P., Canter B., Dynlacht B. D., Primary cilia control hedgehog signaling during muscle differentiation and are deregulated in rhabdomyosarcoma. Proc. Natl. Acad. Sci. U.S.A. 111, 9151–9156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendjan S., Mascetti V. L., Ortmann D., Ortiz M., Karjosukarso D. W., Ng Y., Moreau T., Pedersen R. A., NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310–325 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Tripathi S., Miyake T., McDermott J. C., Smad7:β-catenin complex regulates myogenic gene transcription. Cell Death Dis. 10, 387–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Z., Wang L., Bartom E., Marshall S., Rendleman E., Ryan C., Shilati A., Savas J., Chandel N., Shilatifard A., β-Catenin/Tcf7l2–dependent transcriptional regulation of GLUT1 gene expression by Zic family proteins in colon cancer. Sci. Adv. 5, eaax0698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim L. S., Hong F. H., Kunarso G., Stanton L. W., The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells 28, 1961–1969 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., von Mering C., STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tosic J., Kim G.-J., Pavlovic M., Schröder C. M., Mersiowsky S.-L., Barg M., Hofherr A., Probst S., Köttgen M., Hein L., Arnold S. J., Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 21, 1518–1531 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Fujimi T. J., Hatayama M., Aruga J., Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/β-catenin signaling pathway. Dev. Biol. 361, 220–231 (2012). [DOI] [PubMed] [Google Scholar]
- 44.G. C. Schoenwolf, S. B. Bleyl, P. R. Brauer, P. H. Francis-West, Larsen’s Human Embryology (Churchill Livingstone/Elsevier, ed. 5, 2009). [Google Scholar]
- 45.Alev C., Wu Y., Kasukawa T., Jakt L. M., Ueda H. R., Sheng G., Transcriptomic landscape of the primitive streak. Development 137, 2863–2874 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Smith A. S. T., Passey S. L., Martin N. R. W., Player D. J., Mudera V., Greensmith L., Lewis M. P., Creating interactions between tissue-engineered skeletal muscle and the peripheral nervous system. Cells Tissues Organs 202, 143–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon: Fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521–1540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014); http://www.R-project.org/.
- 52.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma’ayan A., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laing A. F., Lowell S., Brickman J. M., Gro/TLE enables embryonic stem cell differentiation by repressing pluripotent gene expression. Dev. Biol. 397, 56–66 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Mulas C., Chia G., Jones K. A., Hodgson A. C., Stirparo G. G., Nichols J., Oct4 regulates the embryonic axis and coordinates exit from pluripotency and germ layer specification in the mouse embryo. Development 145, dev159103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buscarlet M., Stifani S., The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17, 353–361 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/12/eabf7412/DC1


