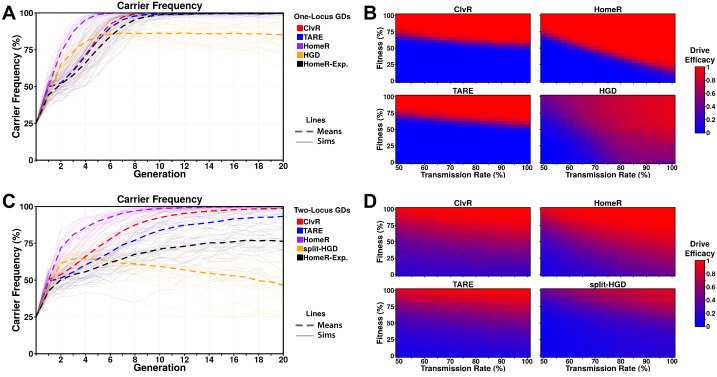
Figure 3. Performance of contemporary gene drive systems for population modification with a single release.
(A) Simulations of carrier frequency trajectories (i.e. heterozygotes and homozygotes) for one-locus versions of ClvR, TARE, HomeR, and HGD for ideal parameters (see 'Materials and methods'), and HomeR for experimental parameters (HomeR-Exp, see 'Materials and methods'). Twenty-five repetitions (lighter lines) were used to calculate the average behavior of each drive (thicker, dashed lines). Populations were initialized with 50% wildtype (+/+) adult females, 25% wildtype (+/+) adult males, and 25% drive homozygous (drive/drive) males. (B) Heatmaps depicting drive efficacy for one-locus versions of ClvR, TARE, HomeR, and HGD for a range of fitness and transmission rate parameter values. Fitness costs were incorporated as a dominant, female-specific fecundity reduction. Transmission rate was varied based on cleavage rate, using HDR rates consistent with ideal parameters, when applicable (see 'Materials and methods'). Drive efficacy is defined as the average carrier frequency at generation 20 (approximately 1 year, given a generation period of 2-3 weeks) based on 100 stochastic simulations with the same initial conditions as (A). (C) Simulations of carrier frequency trajectories for two-locus (split-drive) versions of ClvR, TARE, HomeR, and HGD for ideal parameters (see 'Materials and methods'), and HomeR for experimental parameters (HomeR-Exp, see 'Materials and methods'). Twenty-five repetitions (lighter lines) were used to calculate the average behavior of each drive (thicker, dashed lines). Populations were initialized with 50% wildtype (+/+; +/+) adult females, 25% wildtype (+/+; +/+) males, and 25% drive homozygous (Cas9/Cas9; gRNA/gRNA) males. (D) Heatmaps depicting drive efficacy for two-locus versions of ClvR, TARE, HomeR, and HGD for a range of fitness and transmission rate parameter values, implemented as in panel (B), with initial conditions given in (C).