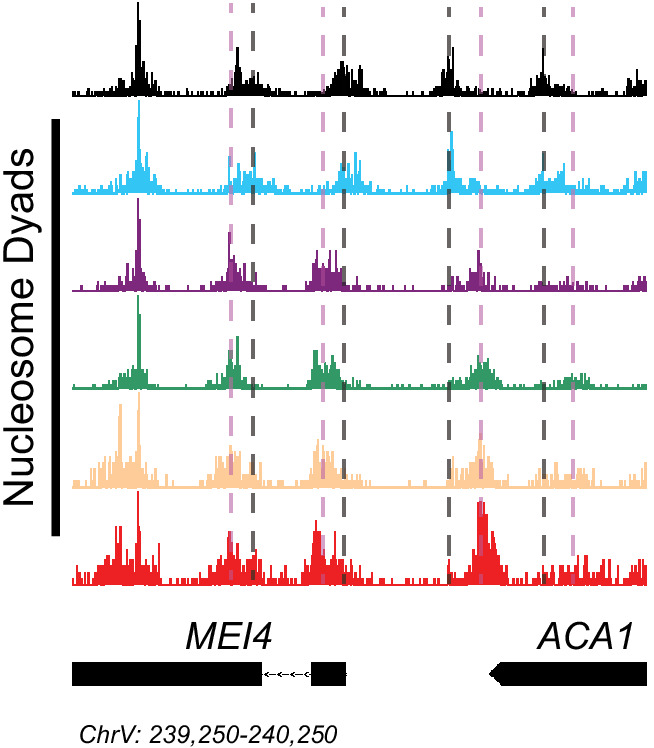
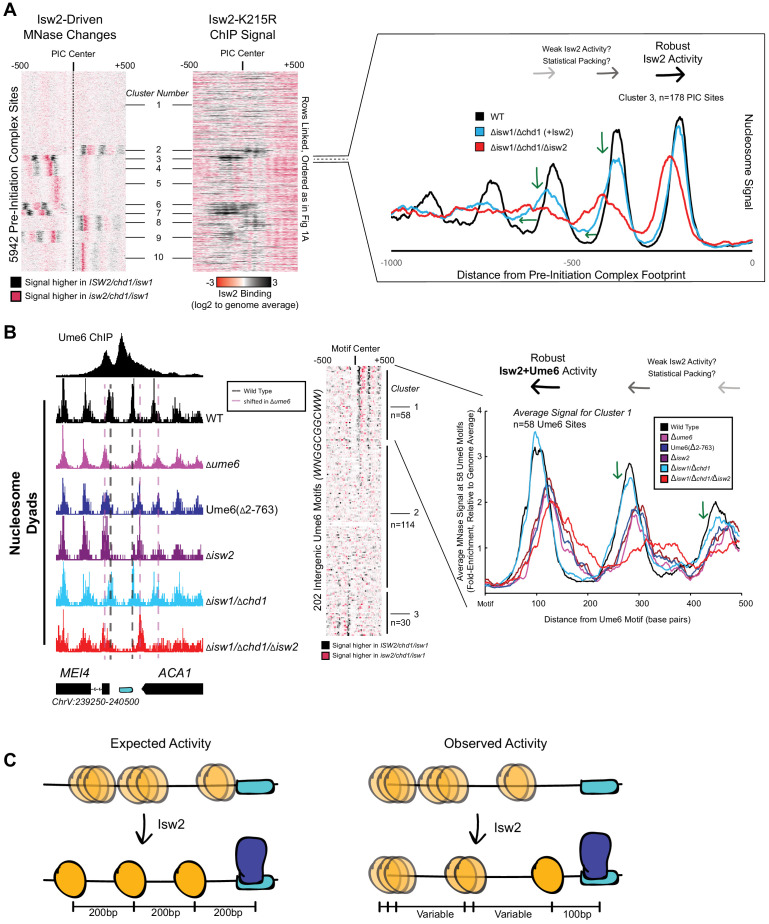
Figure 1. Isw2 is a specialist remodeler that positions single nucleosomes at target sites.
(A) (Left) Clustered heatmap showing differences in nucleosome dyad signal between isw2/isw1/chd1 and ISW2/isw1/chd1 strains at 5942 pre-initiation complex sites (PICs). Black indicates positions where Isw2 preferentially positions nucleosomes compared to the strain lacking Isw2. (Middle) Heatmap of ISW2(K215R) ChIP signal, with rows linked to the PIC data on the left, shows that Isw2-dependent nucleosome changes overlap with regions where Isw2 is present. (Right) Average nucleosome dyad signal for wild type (WT) (black), isw1/chd1 (cyan), and isw2/isw1/chd1 (red) strains for the 178 PIC sites in cluster 3. Black arrows denote Isw2-driven nucleosome shifts. Green arrows indicate rapid decay of positioning at PIC-distal nucleosomes in the ISW2/isw1/chd1 mutant. (B) (Left) Genome Browser image showing nucleosome dyad signal at a unscheduled meiotic gene expression (Ume6) motif (cyan rectangle) for indicated strains. Vertical gray dashed line denotes the motif-proximal WT nucleosome positions while vertical pink dashed line indicates the nucleosome positions in the absence of Ume6 or Isw2. (Center) Clustered heatmap showing the difference in nucleosome dyad signal between isw2/isw1/chd1 and ISW2/isw1/chd1 strains at 202 intergenic Ume6 motifs. Black indicates positions where Isw2 preferentially positions nucleosomes compared to strains lacking Isw2. (Right) Average nucleosome dyad signal for indicated strains at Ume6 motifs in cluster 1. Black arrows indicate direction of nucleosome positioning by Isw2. Green arrows signify decreased positioning of motif-distal nucleosomes in the ISW2/isw1/chd1 strain (cyan) compared to WT (black). (C) (Left) Cartoon depicting the expected activity of Isw2 at barrier elements according to current biochemical data and nucleosome positioning models. Isw2 is thought to move nucleosomes away from bound factors and space nucleosomes with an approximately 200 base pair repeat length. (Right) Cartoon of the observed activity of Isw2 at target sites where only a motif-proximal single nucleosome is precisely positioned but distal nucleosomes are not well-spaced by Isw2.
Figure 1—figure supplement 1. Isw2 is a precise specialist at target nucleosomes.
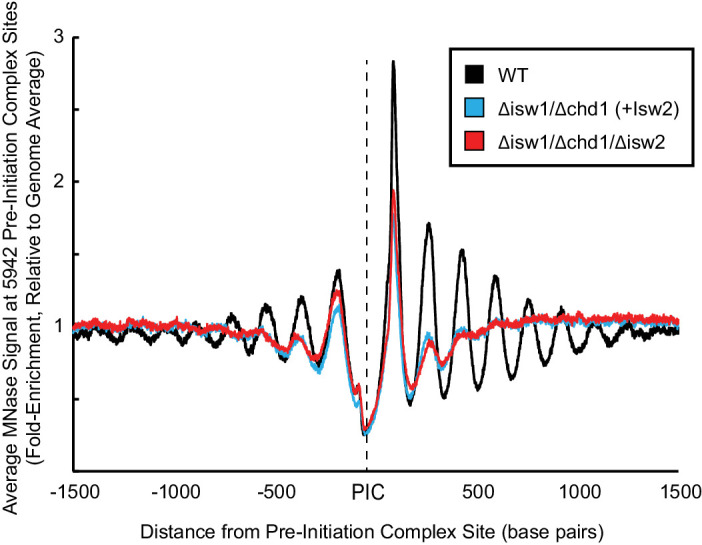
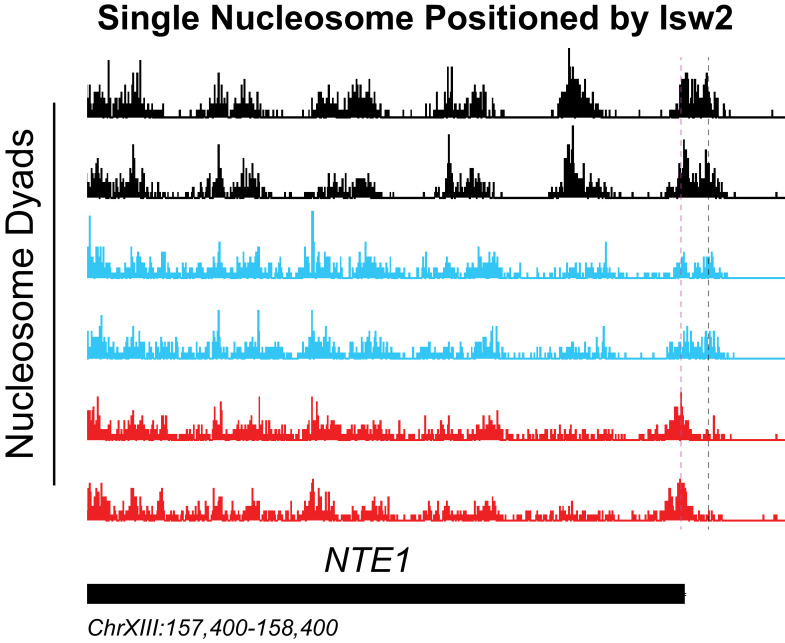
Figure 1—figure supplement 2. Isw2 is a precise specialist at target nucleosomes.
Figure 1—figure supplement 3. Isw2 is a precise specialist at target nucleosomes.
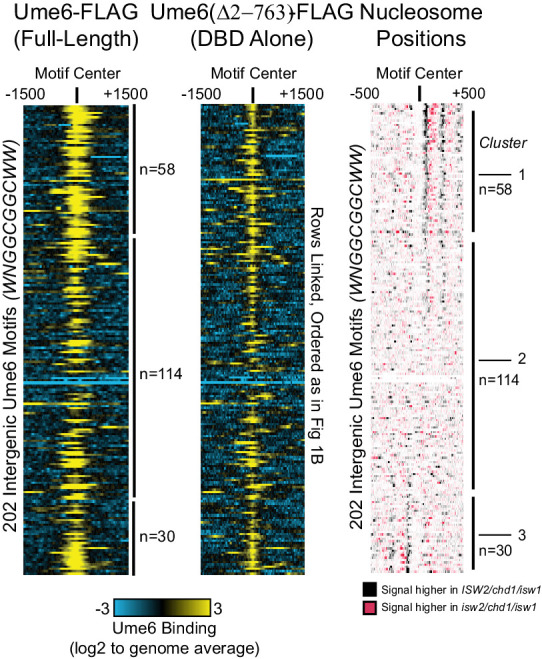
Figure 1—figure supplement 4. Isw2 is a precise specialist at target nucleosomes.
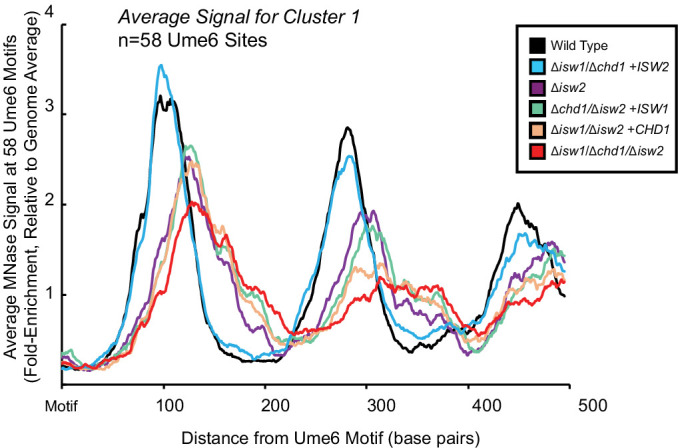
Figure 1—figure supplement 5. Isw2 is a precise specialist at target nucleosomes.