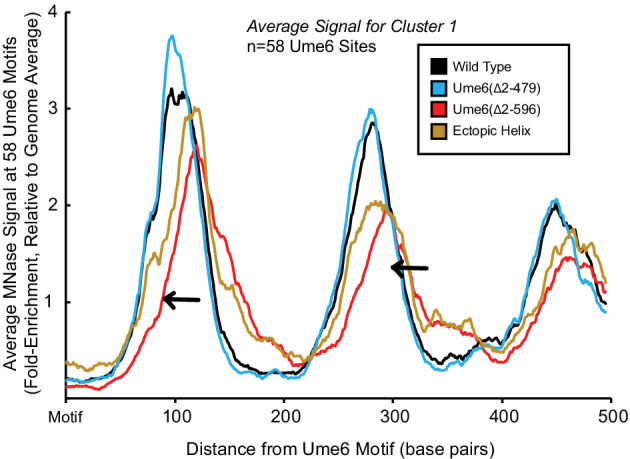
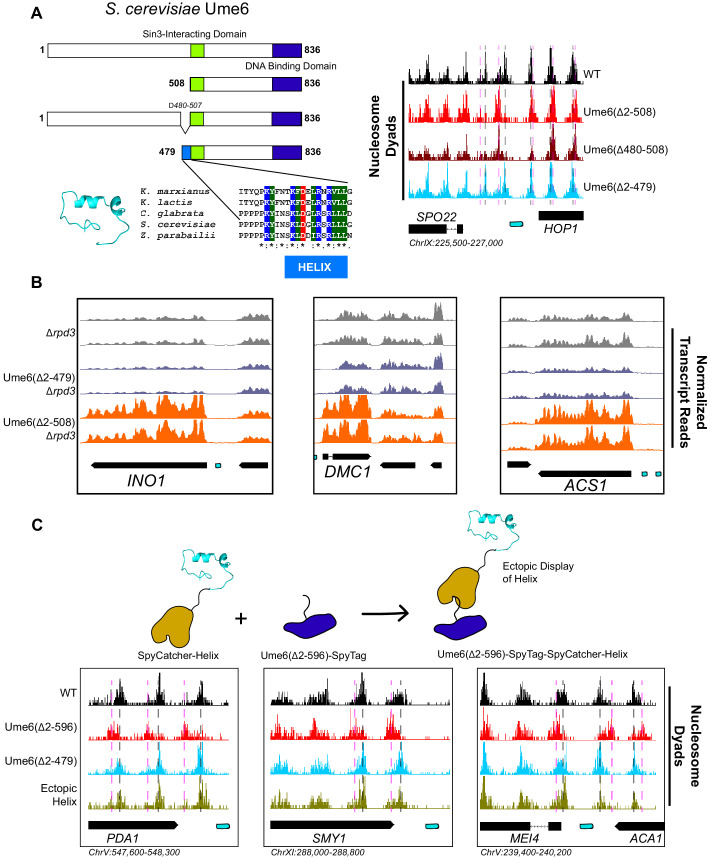
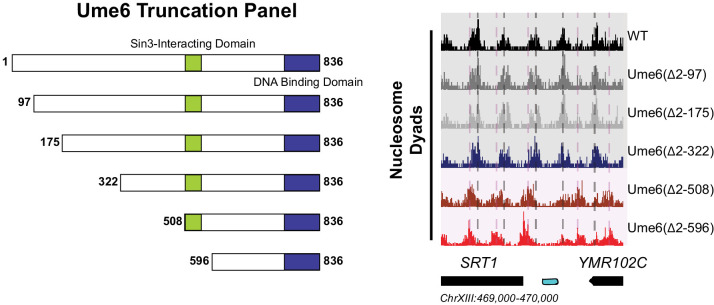
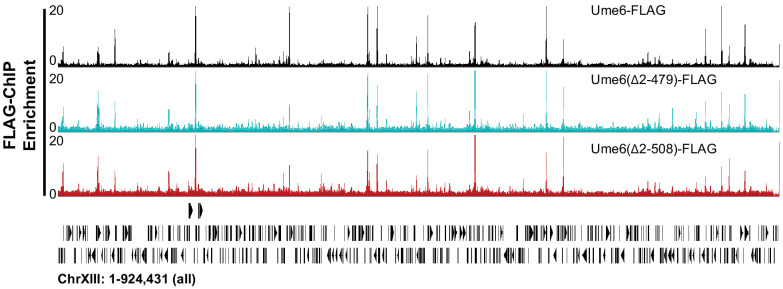
Figure 2. A small predicted helix is the Isw2-recruitment epitope in unscheduled meiotic gene expression (Ume6).
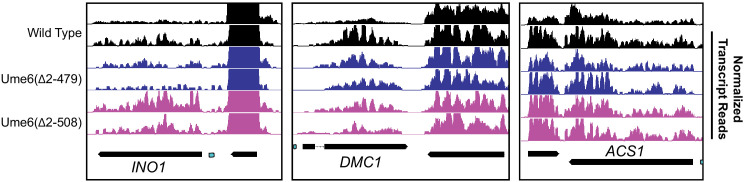
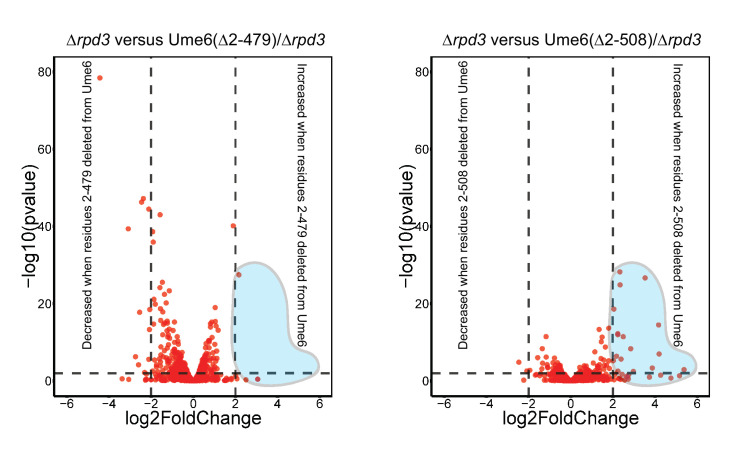
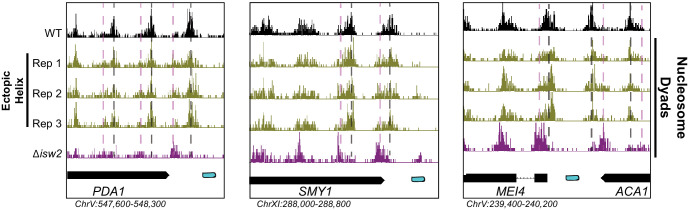
(A) (Top left) Schematic diagram of Ume6 truncation and deletion constructs used to identify the Isw2-recruitment epitope, with the known Sin3-interacting domain depicted as a green square, the DNA binding domain as a dark blue rectangle, and the putative Isw2-recruitment helix as a light blue rectangle. (Bottom left) Modeled helical peptide (by Phyre2) and sequence conservation of the identified Isw2-recruitment motif in Ume6 constructs from other yeasts. Asterisks denote invariant residues. (Right) Nucleosome dyad signal for Ume6 truncation and deletion strains indicates deletion of the region from residues 480 to 507 completely abrogates nucleosome positioning by Isw2 at Ume6 target sites. Vertical dashed gray lines denote wild-type (WT) positions of nucleosomes while vertical dashed pink lines indicate isw2 or ume6-deficient positions of nucleosomes. (B) Genome Browser image showing transcript abundance at three Ume6 target sites for yeast strains lacking Rpd3 with WT Ume6 (gray), Ume6(∆2–479) (blue), and Ume6(∆2–508) (orange). Grossly increased transcription is seen when residues 480–507 are deleted, consistent with expected transcriptional increase associated with loss of Isw2 and Rpd3. Upstream repression sequence (URS) sites are indicated as cyan rectangles. No significant increase in transcription is detected when Ume6 residues 2–479 are deleted. Biological replicates are shown to highlight reproducibility. (C) (Top) Cartoon schematic for ectopic display of the Isw2-recruiting helix (residues 480–507) to the C-terminus of a truncated Ume6 construct lacking Isw2-directed nucleosome positioning. A short SpyTag is appended to the C-terminus of the Ume6 construct and residues 480–507 are fused to the SpyCatcher domain and introduced on a yeast expression vector. (Bottom) Nucleosome dyad signal demonstrating recovery of Isw2-directed nucleosome positions at a subset of Ume6 target genes by the ectopically displayed helical element. Vertical dashed gray lines denote WT positions of nucleosomes while vertical dashed pink lines indicate isw2 or ume6-deficient positions of nucleosomes. URS sites are indicated as cyan rectangles. Individual biological replicates for nucleosome positions after ectopic display of the recruitment helix are provided in Figure 2—figure supplement 4.
Figure 2—figure supplement 1. The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
Figure 2—figure supplement 2. The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
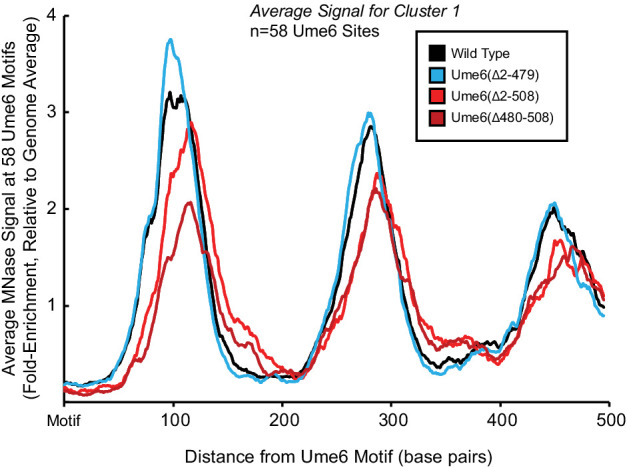
Figure 2—figure supplement 3. The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
Figure 2—figure supplement 4. Transcription data support a role of unscheduled meiotic gene expression (Ume6) residues 479–508 for Isw2 recruitment and not Rpd3 activity.
Figure 2—figure supplement 5. Transcription data support a role of unscheduled meiotic gene expression (Ume6) residues 479–508 for Isw2 recruitment and not Rpd3 activity.
Figure 2—figure supplement 6. Ectopic display of the unscheduled meiotic gene expression (Ume6) helical element can rescue Isw2 activity at Ume6 targets.