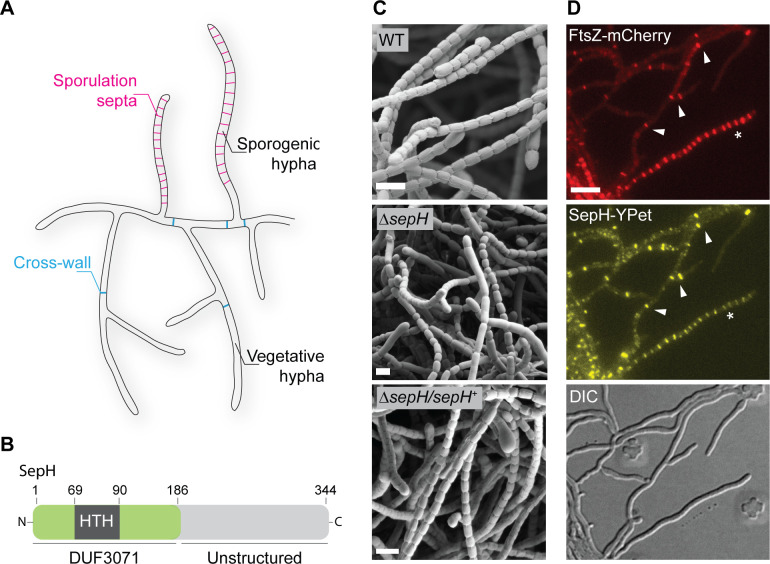
Figure 1. sepH is required for sporulation septation in Streptomyces venezuelae.
(A) Schematic illustrating the multicellular life style of Streptomyces including the two FtsZ-dependent modes of cell division that occur in vegetative and sporogenic hyphae: cross-wall formation and sporulation septation. (B) Schematic of the predicted SepH domain organization including the N-terminal DUF3071 domain containing a helix-turn-helix (HTH) motif and the unstructured C-terminal domain. Numbers indicate corresponding amino acid positions. (C) Cryo-scanning electron micrographs of sporogenic hyphae from wild-type (WT) S. venezuelae, the ΔsepH mutant (SV56), and the complemented mutant strain ΔsepH/sepH+ (MB747). Scale bars: 2 μm. (D) Subcellular co-localization of fluorescently labeled FtsZ (FtsZ-mCherry) and SepH (SepH-YPet) in vegetative and sporulating hyphae. Fluorescent gene fusions were expressed in the WT background (MB751). White arrow heads point at co-localization at cross-walls in vegetative hyphae and the asterisk denotes a sporogenic hypha undergoing sporulation septation. Scale bar: 5 µm.
Figure 1—figure supplement 1. sepH is a direct target of the transcriptional regulators WhiA and WhiB.

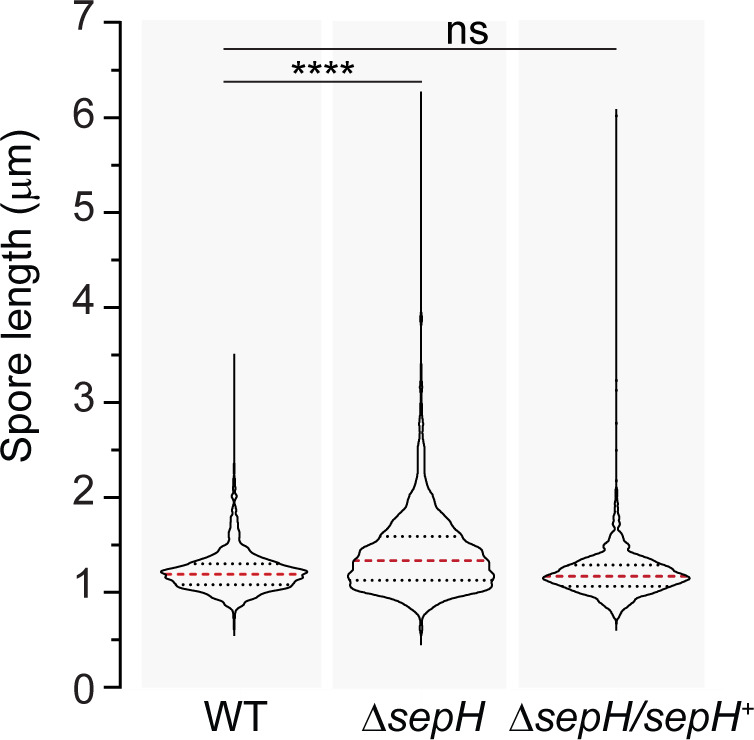
Figure 1—figure supplement 2. Spore length analysis of wild-type (WT) S. venezuelae, the ΔsepH mutant (SV56), and the complemented mutant strain ΔsepH/sepH+ (MB747).