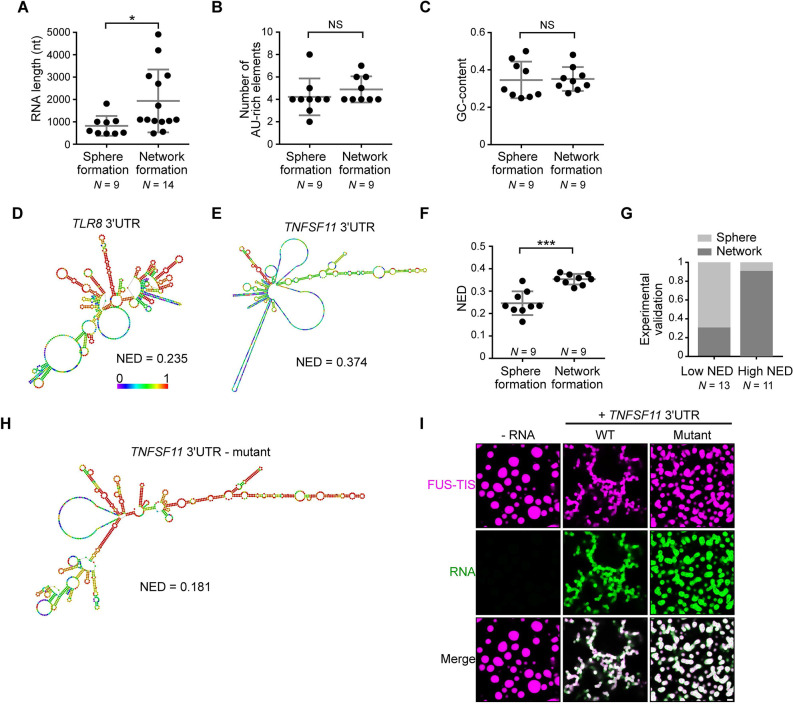
Figure 3. RNAs predicted to have large disordered regions have a high propensity to induce network formation in vitro.
(A) Distribution of length of sphere- and network-forming RNAs. Mann–Whitney test, Z = −2.76, p=0.004. See also Figure 3—source data 1. (B) Number of AU-rich elements in sphere- and network-forming RNAs with a length shorter than 2000 nt. See also Figure 3—source data 1. Mann–Whitney test, Z = 0.190, p=0.258, NS, not significant. (C) Distribution of GC-content of sphere- and network-forming RNAs with a length shorter than 2000 nt. See also Figure 3—source data 1. Mann–Whitney test, Z = 0.566, p=0.605. (D) Centroid RNA secondary structure of TLR8 3′UTR predicted by RNAfold. The color code represents base-pairing probability. (E) Same as (D), but the TNFSF11 3′UTR is shown. (F) Normalized ensemble diversity (NED) values of sphere- and network-forming RNAs. See Figure 3—source data 1. Mann–Whitney test, Z = −3.3, ***p<0.0003. (G) Experimental validation of N = 24 in vitro transcribed RNAs whose ability for network formation was predicted by NED. Sphere formation is indicated in dark gray, whereas network formation is indicated in light gray. See Figure 3—source data 1. Mann–Whitney test was performed on the experimental validation, Z = −2.8, ***p=0.004. (H) Same as (D), but the mutant TNFSF11 3′UTR is shown. (I) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of 150 nM of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm.