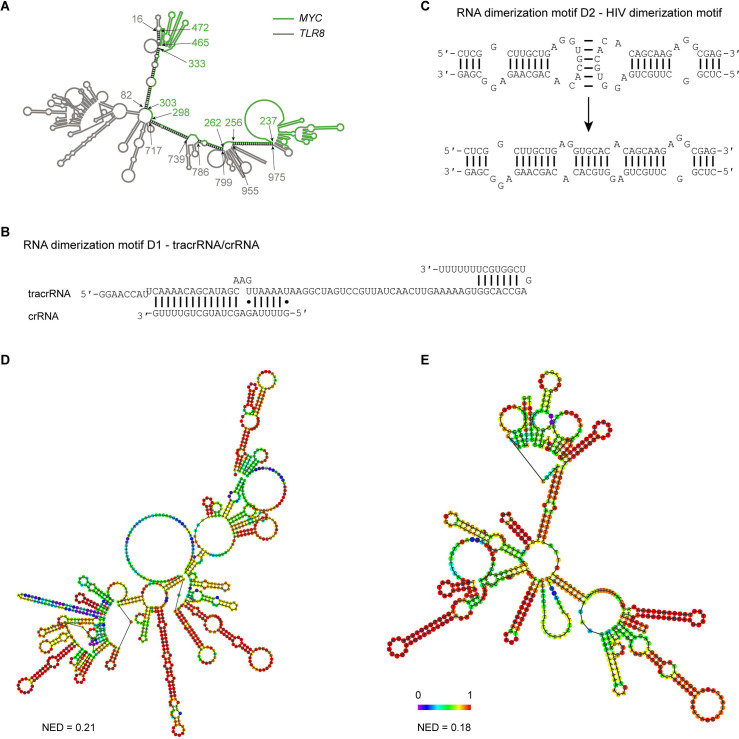
Figure 5. A multivalent RNA matrix is responsible for mesh-like condensate formation in vitro.
(A) Schematic of RNAs with strong local secondary structures that are predicted to induce spherical condensates. (B) Schematic of RNAs with large disordered regions that form extensive intermolecular RNA–RNA interactions that are predicted to form network-like condensates. (C) Native and denaturing agarose gel electrophoresis of sphere-forming (lanes 1–3) and network-forming (lanes 4–6) RNAs (5 µM, each [1.7, 1.6, 1.9, 1.7, 1.8, 1.7 µg/µl]). (D) Schematic of a complex RNA network characterized by extensive intermolecular RNA–RNA interactions mediated by two dimerization elements (D1 and D2) that were added to structured RNAs. (E) Native and denaturing agarose gel electrophoresis of the indicated RNAs (1 µM, each [326, 154, 345, 193 ng/µl]). (F) Representative images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of the indicated Cy3- or Cy5-labeled RNAs generated by in vitro transcription after 16 hr of incubation. Scale bar, 2 µm.