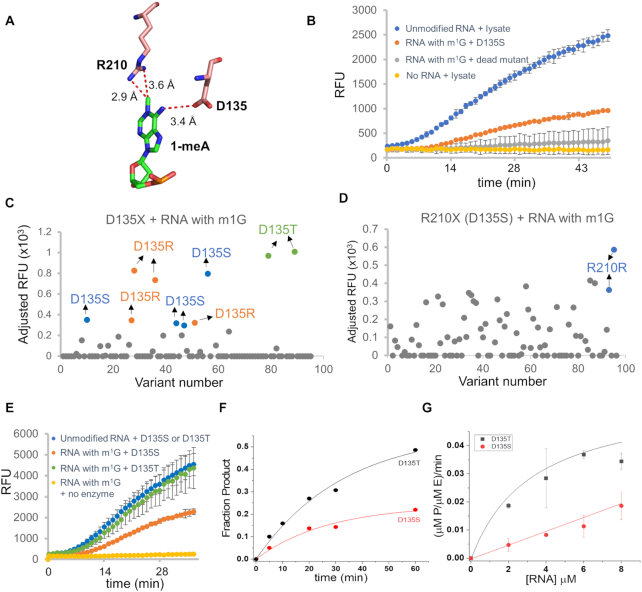
Figure 2.
Screening at positions D135 and R210 of AlkB. (A) Residues D135 and R210 are located in close proximity to the modified nucleobase in the crystal structure (3BIE) (16). (B) Under the optimized condition, a high dynamic range was achieved to allow for the identification of mutants with higher reactivity. Error bar indicates SD, n≥3. (C, D) Screening results for libraries D135X and R210X (D135S). Each dot represents one variant in the library. The absolute fluorescence signal generated with each member in the library was adjusted by subtracting it from the signal generated with the dead mutant. Variants with adjusted signals below zero were presented as zero to indicate that no activity was observed for these variants under the condition. (E) In vitro demethylation assay with purified D135T and D135S mutants using the fluorescence assay confirmed that D135T has higher activity than D135S to demethylate m1G. The demethylation activity was judged by comparing with the signal of the positive control, which was the unmodified RNA incubated with the respective variant. For direct comparison between the two variants, data were adjusted so that the positive controls for the two variants had the same level of fluorescence signals. (F) Plot of fraction of product as a function of time for the reaction containing 4 μM enzyme and 4 μM 9-mer m1G-RNA. (G) Plot of (μM P/μM E)/min as a function of RNA concentration for reactions containing 4 μM enzyme. The value of (μM P/μM E)/min was calculated as ([S]/[E])*A*k (see Materials and Methods). Data shown are the average and standard deviation of three independent reactions for each condition.