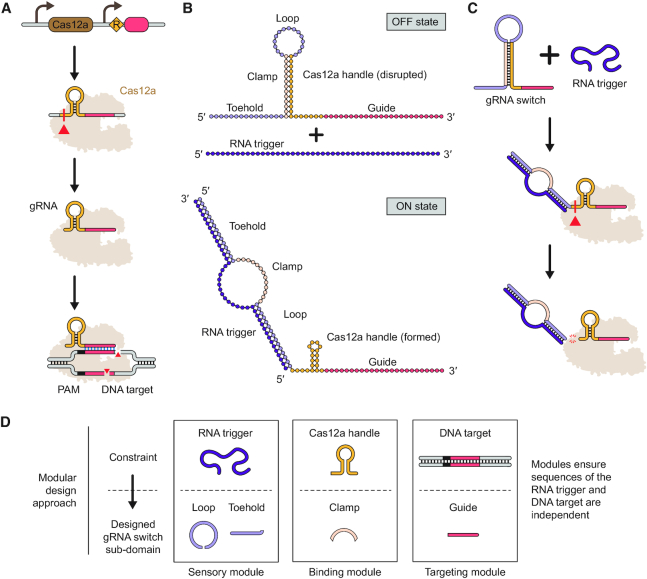
Figure 1.
Modular design of Cas12a gRNA switches with unconstrained RNA triggers and DNA targets. (A) gRNA processing and DNA targeting by Cas12a. The gRNA consists of a direct repeat and spacer. The repeat forms a canonical hairpin ‘handle’ recognized by Cas12a, which cleaves upstream of the hairpin. The downstream portion of the repeat and the spacer are retained as the processed gRNA utilized by Cas12a for targeted cleavage of complementary DNA sequences flanked by a PAM. (B) Architecture and intended conformational states of the gRNA switch. In the absence of an RNA trigger (OFF state), the clamp base pairs with the Cas12a handle, disrupting its formation and subsequent recognition by Cas12a. In the presence of an RNA trigger (ON state), the toehold and loop base pair with the RNA trigger, leading to an energetically favorable conformation wherein the Cas12a handle re-folds and can be recognized by Cas12a. The split sensor domain does not overlap with the clamp or the guide sequences, resulting in sequence independence of the trigger, Cas12a handle and guide sequences. Both conformational states as well as the desired guide are supplied to NUPACK to design the trigger, toehold and loop. (C) Switch activation and processing in the presence of an RNA trigger. Hybridization between the RNA trigger and the gRNA switch drives a conformational change that allows formation of the Cas12a handle. Handle recognition leads to gRNA switch processing via Cas12a's inherent endoribonuclease activity. (D) The modular design approach for gRNA switches. Each module links a design constraint with a corresponding switch sub-domain.