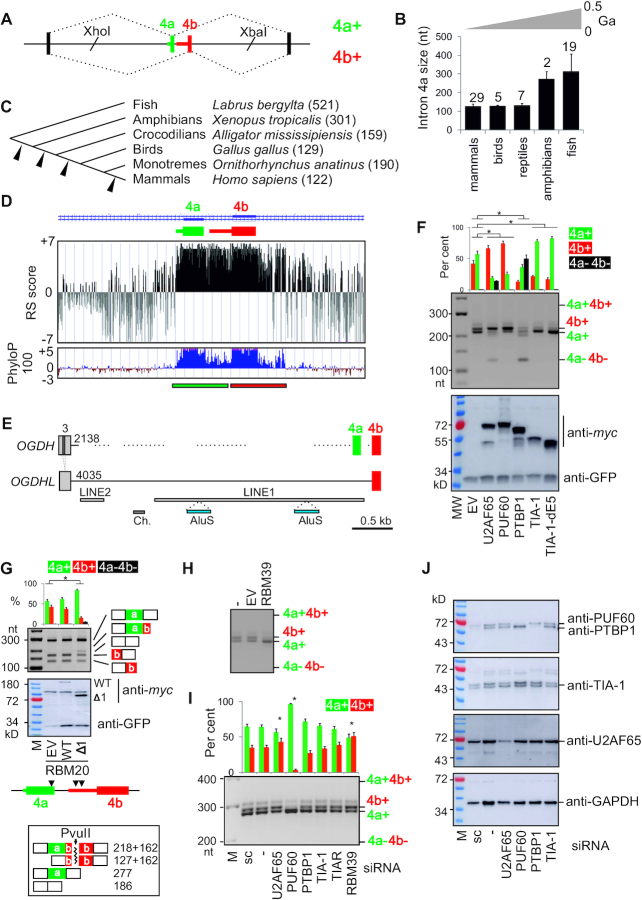
Figure 1.
Intron 4a as a regulator of OGDH MXEs and target for pyrimidine-binding proteins that control Ca2+-sensitive and -insensitive isoforms. (A) OGDH reporter construct. Introns are shown as horizontal lines, exons as boxes and spliced products (right) by dotted lines. MXEs are numbered at the top. Introns are numbered by their genomic location in the main text. The length of MXE PPTs is indicated by coloured horizontal bars. (B) Average size of intron 4a in vertebrate classes. Error bars are SDs. The number of species is indicated above each bar. Approximate evolutionary span is at the top (not to scale). Ga, giga-annum. (C) Intron 4a size in representative species (in nts). Arrowheads denote deletion events. (D) RS (rejected substitutions) scores (65) (upper panel) and PhyloP 100 vertebrates conservation (lower panel) of the MXE pair. Rectangles at the bottom denote a tentative exon duplication event. (E) TEs (horizontal bars) in OGDH and OGDHL introns 3 and 4a. Their size (in nts) in the reference genome is shown at their 5′ ends. Exons are numbered at the top. Dotted lines show regions weakly aligned with OGDHL. LINE, long interspersed elements; Ch., DNA transposon Charlie. (F) Regulation of OGDH MXEs by U-binding proteins. The OGDH reporter was cotransfected with expression constructs (bottom). OGDH mRNA isoforms (middle panel) are measured at the top. Error bars are SDs of two independent transfections. Asterisks denote significant P values (<0.05; ANOVA, Dunnett's post-hoc tests) for the indicated comparisons. MW, size marker; EV, empty vector. Immunoblot with lysates prepared from transfected cells is in the lower panel; antibodies are to the right. (G) RBM20 is a repressor of OGDH exon 4b. Here, RT-PCR products are cut with PvuII, which cleaves exon 4b; digested products and their sizes (in nts) are shown at the bottom. Coexpression of the OGDH reporter with RBM20 constructs is shown in the middle panel; arrowheads below denote the presence of optimal binding sites of RBM20 (UCUU) (81) located in the 5′ part of megaPPT (segment del2, Figure 2A) and in the 3′ part of exon 4a. Ogdh was identified among cardiomyocyte RBM20 HITS-CLIP targets (10 intronic reads) although no Ogdh exons were listed among high-confidence RBM20-regulated exons (FDR<0.05) identified by comparing Rbm20 (−/−) and WT rats (81). (H, I) Opposite effects of RBM39 overexpression (H) and depletion (I, last lane) (40) on exon 4a/4b ratios. (I) Exon 4a/4b usage in HEK293 cells depleted of Y-binding proteins. sc, a scrambled control. Spliced products are measured at the top; error bars are SDs of two transfections. Asterisks show statistically significant deviations from controls (ANOVA with post-hoc Dunnett's tests). (J) Immunoblots for panel (I); antibodies are to the right.