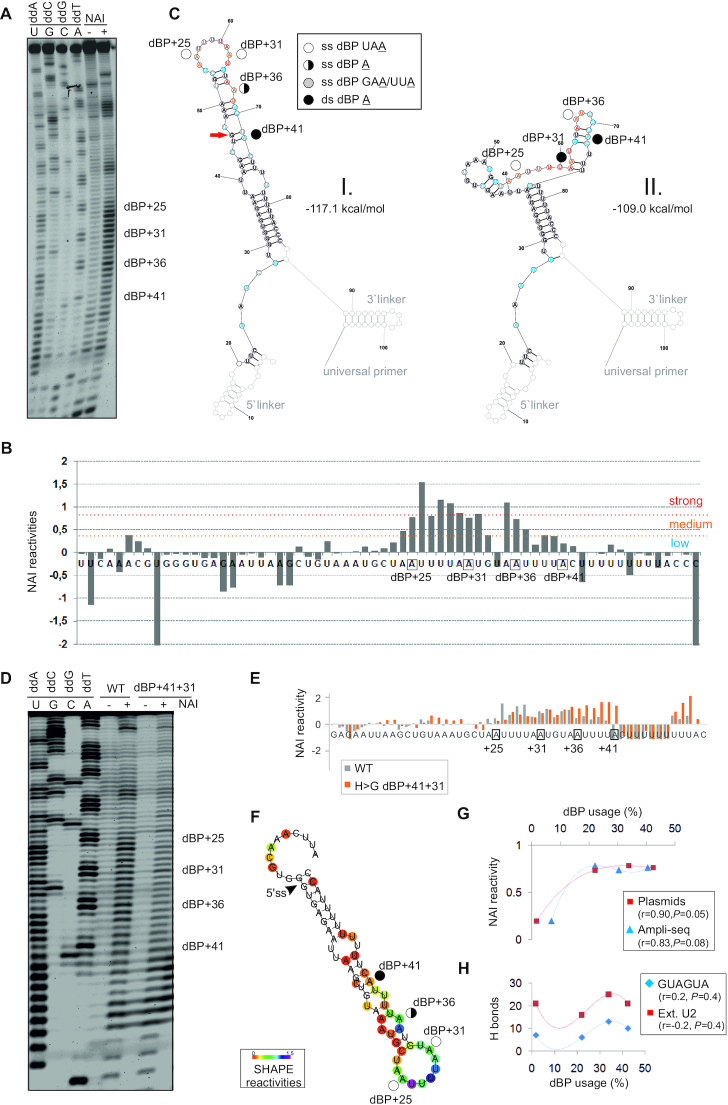
Figure 8.
SHAPE with human RNA probes modified by NAI. (A) Separated RT products of in vitro NAI-treated (+) or untreated (−) reactions with human Cy5-labeled WT RNA probe with linkers. dBP cluster is labeled to the right. Dideoxy sequencing ladder (lanes 1–4) was run in parallel to determine modified positions. (B) Normalized NAI reactivities. Nucleotide flexibility is coloured (scale is to the right); negative values are shown in full. (C) The most stable SHAPE-guided structure (I, left) with an alternative stable structure (II, right). Predictions were carried out with RNAstructure using reactivity values from panel B. Linkers are greyed. Red arrow shows a wobble pair at the end of helix; if disallowed in RNAfold (59), dBP+41becomes single-stranded. (D) Human WT and mutated probes that lack linkers. (E) Normalized NAI reactivities for panel D; negative values are cut off at −1. (F) Optimal SHAPE-guided secondary structure of the WT RNA probe supported by methods developed by Deigan et al. (55) (default slope m = 1.9 and intercept b = −0.7), Washietl et al. (57) (linear mapping and tau/sigma-ratio of 1) and Zarringhalam et al. (56) (linear linear log model, slope 1.6, intercept −2.29 and β = 0.8). (G,H) Usage of individual dBPs correlates with their NAI reactivities (G) rather than predicted U2:BP base-pairing (H), shown as the number of hydrogen bonds between dBPs and canonical GUAGUA motif (blue) or the extended U2 snRNA region (red). r, correlation coefficients and associated P values (F-tests). For Ampli-seq data, r and P values were 0.65 and 0.18 (GUAGUA) and 0.26 and 0.4 (extended U2 motif), respectively.