Figure 3.
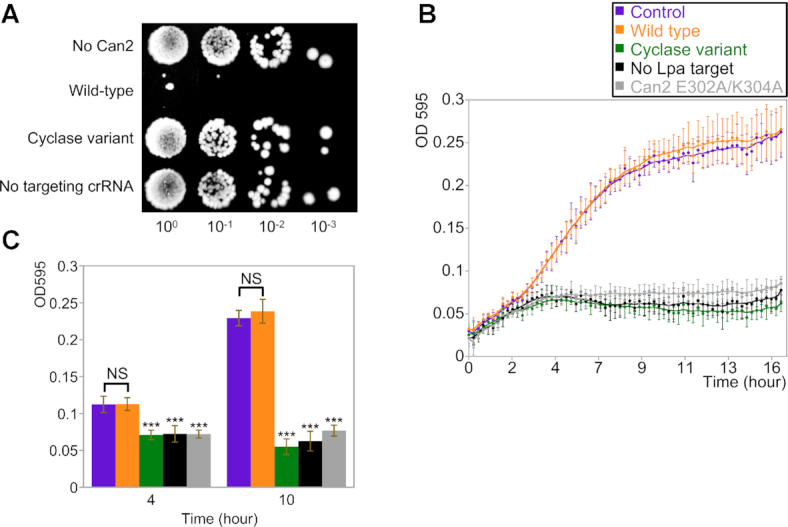
Can2 protects against MGE in vivo. (A) Plasmid challenge assay of mycobacterial type III-A system in E. coli host. E. coli cells harbouring the mycobacterial type III-A interference complex Csm1-5 and TetR targeting spacer were transformed with plasmid containing TsuCan2 effector and tetracycline-resistance gene (Wild-type). Other strains are indicated as ‘No Can2’ where the TsuCan2 is absent, ‘Cyclase variant’ where the interference complex is unable to produce cOA and ‘No targeting crRNA’ where the TetR targeting spacer is replaced with a spacer targeting pUC19 MCS. A 10-fold dilution series of the transformation mixture was applied onto tetracycline selective plates to determine the number of viable transformants. (B) Growth curves of E. coli cells harbouring the interference complex and phage P1 lpa gene-targeting spacer supplemented with TsuCan2 effector (indicated as Wild-type). Cells were grown in LB broth in a 96-well plate with shaking at 37°C and infected with phage P1 at a MOI of ∼1. OD595 of the culture was measured every 15 min to plot against time over the 16 h incubation. Other strains are indicated as ‘Cyclase variant’ (green) where the interference complex is unable to produce cOA molecules; ‘No Lpa target’ (black) where lpa gene-targeting spacer is replaced with a spacer targeting pUC19 (MCS); ‘Can2 E302A/K304A’ (grey) which is a TsuCan2 nuclease variant. ‘Control’ (purple) represents wild-type cells (orange) incubated without phage infection under the same conditions. Data points represent the mean of eight experimental replicates (four biological replicates with two technical replicates each) with the standard deviation shown. (C) The OD595 values of all strains after 4 and 10 h growth are shown, coloured as in panel B. Statistical analysis was carried out with RStudio using the unpaired Welch two sample test to calculate P-values. NS (not significant) indicates P-values > 0.05 and *** indicates P-values < 1E–05.
