Figure 3.
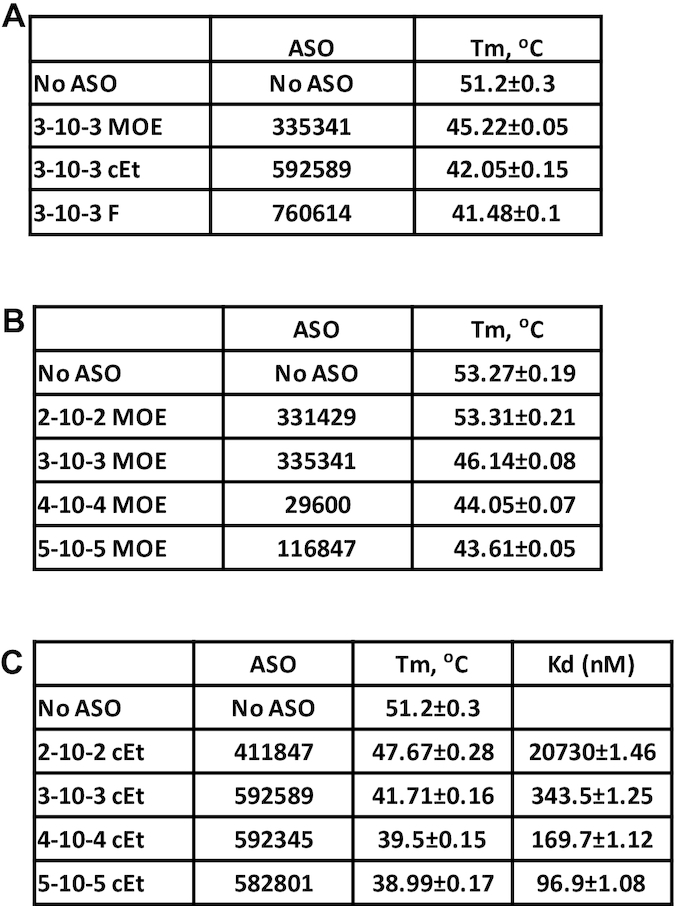
Altering the length of PS-ASOs affects the stability of RNase H1. (A) Thermostability assay was performed to determine the Tm change of H1-NTD upon binding of 3–10–3 gapmer ASOs of the same sequence but with different 2′-modifications. Buffer (Tris-EDTA: 50mM Tris, 100 mM NaCl and 0.5 mM EDTA) is different from Figure 1C, which caused different Tm value. (B, C) H1-NTD stability change upon binding to 2′-MOE (B) or 2′-cEt (C) modified PS-ASOs with different lengths, as determined using PTS assay. Protein binding affinity (Kd, nM) was determined using NanoBRET assay for RNase H1 protein and 2′-cEt modified PS-ASOs of different lengths. The binding Kd (nM) was calculated using Prism. The average values and standard deviations from four duplicates are shown.
