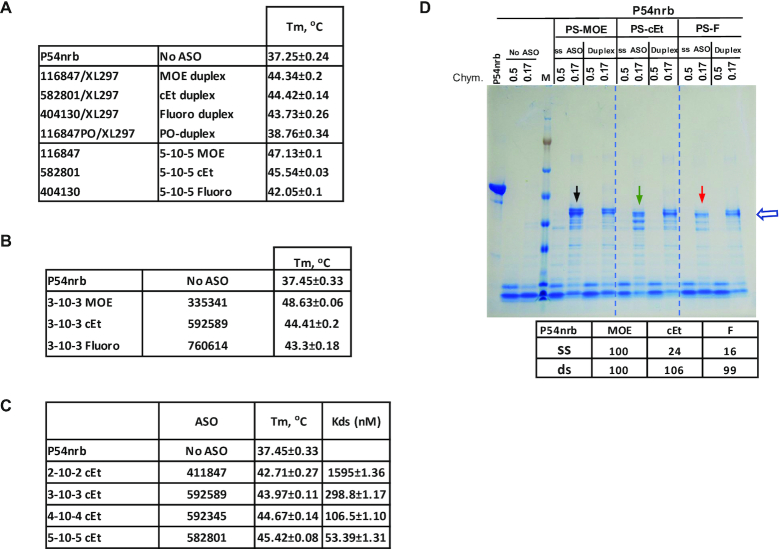
Figure 5.
PS-ASO binding can affect the stability of P54nrb protein and cause conformational change. (A) Thermostability assay was performed to determine the Tm change of P54nrb upon binding of 5–10–5 gapmer ASOs of the same sequence but with different 2′-modifications or with ASO/RNA duplexes formed with a complementary RNA and these PS-ASOs. (B, C) P54nrb protein stability change upon binding to 3–10–3 PS-ASOs with different 2′-modifications (B), or to PS-cEt ASOs with different lengths (C), as determined using PTS assay. Protein binding affinity was determined using NanoBRET assay for P54nrb protein and 2′-cEt modified PS-ASOs of different lengths. The binding Kds (nM) were calculated using Prism. The average values and standard deviations from four duplicates are shown in related panels A, B and C. (D) Coomassie blue staining of P54nrb protein incubated with PS-ASOs or ASO/RNA duplexes, as in panel A, followed by digestion with different concentrations of chymotrypsin. The arrows indicate enhanced digestion by ss-PS-ASOs compared with PS-ASO/RNA duplex. The protein bands marked by the open arrow were quantified and the levels in percentage relative to that in PS-MOE ASO sample are listed.