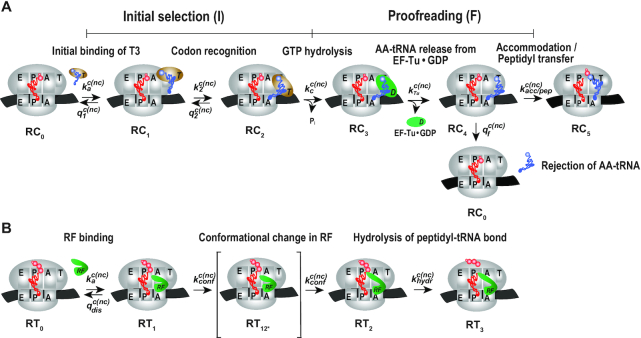
Figure 1.
Kinetic scheme for efficiency and accuracy of codon reading by tRNA (A) and release factors (B). (A) Ribosome complex RC0 has peptidyl-tRNA in P site and A site programmed with native (GAA) or modified (Gm6AA) Glu codon, native (GAU) or modified (Gm6AU) Asp codon. Glu-tRNAGlu, with cognate codon GAA, initially is in free ternary complex (T3) with EF-Tu and GTP. T3 binds into A/T site of RC0, leading to ribosome complex RC1, where the anticodon of T3 lacks codon contact. Codon-anticodon contact is formed by conformational change of tRNA in T3 leading to complex RC2. Subsequent hydrolysis of GTP in EF-Tu induces conformational change of EF-Tu and leads to complex RC3. Then EF-Tu in the GDP form rapidly dissociates from RC3, leading to complex RC4. In RC4 tRNA may be accommodated into A site and receive a nascent peptide from P-site tRNA, leading to complex RC5. Alternatively, tRNA is discarded from the ribosome in a proofreading reaction, which brings RC4 back to complex RC0. The efficiencies (kcat/Km) for GTP hydrolysis on ribosome bound T3 for peptide bond formation in response to GAA, Gm6AA, GAU and Gm6AU codons in A site are in M&M defined in terms of rate constants in Figure 1A (Materials and Methods). (B) Ribosomal termination complex RT0 has peptidyl-tRNA in P site and A site programmed with native (UAA) or modified (Um6AA) stop codon in cognate reactions and with UAG or Um6AG in near-cognate reactions. Release factor 1 (RF1) or 2 (RF2) binds to RT0 forming RT1. Either release factor (RF) undergoes a conformational change and forms complex RT2 via transition state RT12* or dissociates from the ribosome. We suggest that the modest selectivity of ground state RT1 is enhanced in the transition state RT12* (37). Hydrolysis of ester bond in peptidyl-tRNA leads to ribosomal complex RT3 followed by peptide release and, eventually, RF-release. The efficiency (kcat/Km) and maximal rate (kcat) for ester bond hydrolysis on ribosome bound RF in response to UAA, Um6AA, UAG and Um6AG codons are defined in terms of rate constants in Figure 1B (Materials and Methods).