Figure 1.
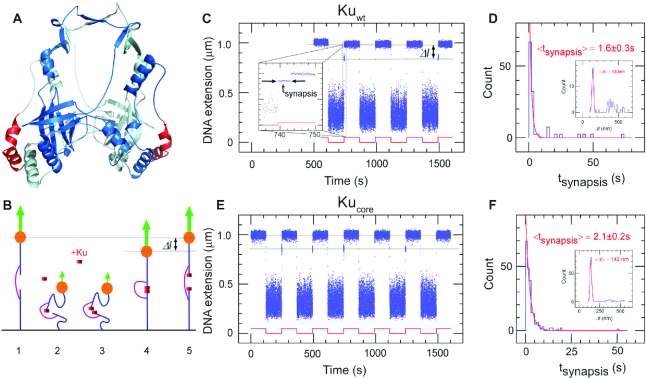
The core structure of bacterial Ku forms short-lived synapsis of blunt DNA ends. (A) 3D model of the bacterial Kuwt homodimer, computed by comparative modelling to the eukaryotic Ku70/80 heterodimer. The red regions highlight the protruding C-terminal arms, which are deleted in the Kucore construct (downstream C-terminal tail of 35 residues predicted as disordered is not represented). (B) Schematic showing the molecular forceps experiment where 1.5 kb (blue) dsDNA segments with blunt ends are joined through a 600 bp dsDNA ‘bridge’ segment (magenta) anchored 59 bases from each end. The blue dsDNA segments are attached to a glass surface (black) and tethered to a magnetic bead (orange), respectively. When extended by a 1.4 pN force (green arrow) by a magnet and in the absence of interacting proteins (1) the DNA construct will have a maximal extension of ∼1 μm. (2) When the force is lowered to the femtoNewton range the ends can meet (3) and for instance be held together by Ku. This can be seen when (4) the DNA extension does not recover to 1 μm when the force is returned to its initial value: in this case the extension of the construct will be shorter by roughly 140 nm. (5) The construct will return to its initial extension upon disruption of the synapse. (C) Representative time-trace obtained upon force modulation (red) in the presence of Kuwt. Inset shows an expanded view of a rupture event of a single end-to-end synapsis. Δl corresponds to the change in DNA extension upon rupture, and tsynapsis the duration of the synaptic event. Events are indexed by coordinate pair (Δl, tsynapsis). (D) Lifetime distribution for end-specific events is fit to a single-exponential distribution (red), yielding a lifetime of 1.6 ± 0.3 s (SEM, n = 51). End-specific events are identified (inset) as having a Δl value within three standard deviations of the mean expected amplitude change given bridge mechanics (Gaussian fit in red, <Δl> = 140 ± 12 nm, SD, n = 99). (E, F), As for C and D but for Kucore. The mean lifetime is derived from a single-exponential fit giving a value of 2.1 ± 0.2 s (SEM, n = 212). Gaussian fit parameters for amplitude distribution are <Δl> = 142 ± 11 nm SD, n = 251).