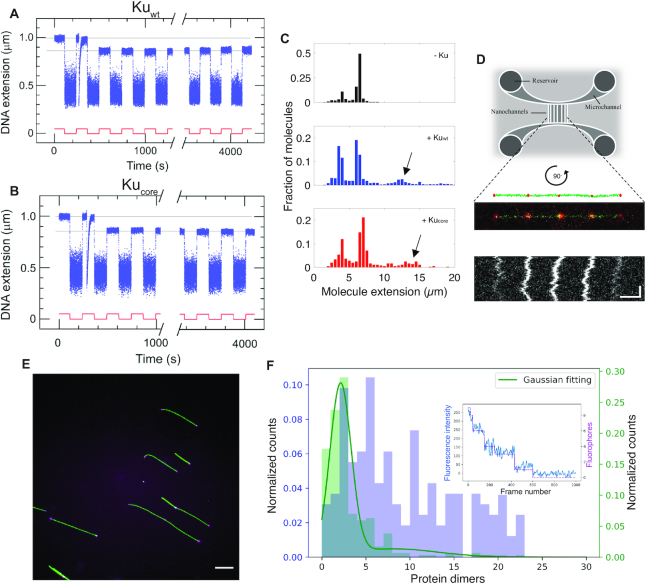
Figure 2.
The core structure of Ku forms long-lived synapsis of sticky DNA ends. Representative time-trace of molecular forceps experiment where a sticky-ended DNA construct with 4 nt complementary overhangs interacts with (A) Kuwt and (B) Kucore. A highly stable synapsis forms, which is not disrupted within 1 h of force-cycling at 1.4 pN. (C) Extension histogram of λ-DNA (black, top, N = 407), λ-DNA + Kuwt (blue, center, N = 1145) and λ-DNA + Kucore (red, bottom, N = 349) in 100 × 150 nm2 nanofluidic channels. Upon addition of protein at a ratio of 100 homodimers per DNA end, circularization and concatemerization of DNA is observed for both Ku variants. The peaks at ∼4 μm and ∼7 μm correspond to single circular and linear λ-DNA molecules, respectively. Longer extensions (indicated by arrows) correspond to concatemers of λ-DNA molecules. (D) Top: A schematic of the nanofluidic chip design, where the vertically aligned nanochannels (100 × 150 nm2) span the two horizontal microchannels. The samples are loaded in the circular reservoirs. Center: A schematic of four λ-DNA molecules (green) joined together by KuAF647 (red) confined in a nanochannel is shown together with the fluorescence microscopy image of a corresponding DNA–protein complex. Bottom: A kymograph shows the positions of the fluorescent KuAF647 over time. Horizontal and vertical scale-bars correspond to 5 μm and 5 s, respectively. (E) Representative fluorescence image of YOYO-1 labelled λ-DNA (green) bound with KuAF647 (magenta) stretched on glass cover slips. Scale-bar corresponds to 10 μm. (F) Normalized histograms for the number of KuAF647 protein dimers bound to λ-DNA (4 μM) at a protein concentration of 33 nM (purple) and a 50 bp blunt-ended DNA substrate (10 nM) in the presence of 120 nM KuAF647 (green). The histograms were constructed from the number of homodimers at different loci on DNA stretched on glass coverslips. The total number of fluorophores on all bound dimers were counted using photobleaching steps analysis. The number of fluorophores was normalized by the protein labeling efficiency (71%) to derive the number of protein dimers. The histogram corresponding to 50 bp DNA was fitted to a Gaussian mixture model (solid green line), from which the total number of homodimers was estimated to be 2.13 ± 1.21. Inset: a representative photobleaching curve (blue) over 1000 frames along with step-detection and enumeration of the number of fluorophores (magenta) A total of 9 fluorophores are observed for this particular λ-DNA-KuAF647 complex.