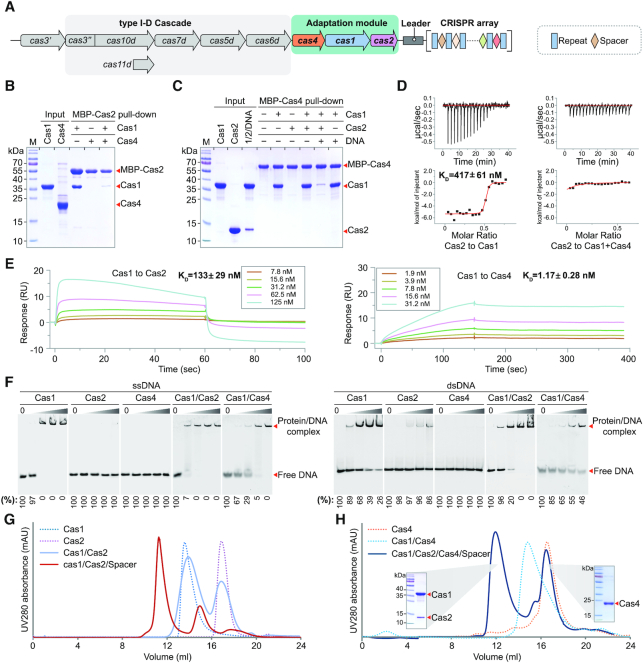
Figure 1.
Reconstitution of the type I-D adaptation module in Synechocystis. (A) Architecture of the genomic locus for the type I-D CRISPR–Cas system from Synechocystis. Spacer, repeat and cas genes are shown as rectangle, diamond and arrow, respectively. (B, C) MBP pull-down assays for assessing interactions between purified Cas1, Cas2 and Cas4. The Cas1–Cas2–prespacer complex is labelled as 1/2/DNA for concision. (D) Binding affinity between Cas1 and Cas2, and between Cas2 and Cas4–Cas1 complex measured by ITC. Untagged Cas proteins are used. The baseline-corrected instrumental response is shown in the upper panel, and the integrated data (squares), together with the best fits (solid lines), are shown in the lower panel. Data are representative of three independent experiments. (E) Binding affinity between adaptation Cas proteins measured by SPR. Untagged Cas proteins are used. Cas2 or Cas4 are immobilized on the sensor chip and Cas1 is used as the analyte. The analyte is injected at a series of 2-fold serial dilutions. Data are representative of three independent experiments. (F) DNA binding ability of Ca1, Cas2 and Cas4. EMSA is performed using 5′ Cy3-labeled 36-nt ssDNA or 36-bp dsDNA. Different concentrations of Cas proteins (0, 0.2, 0.4, 1 and 4 μM) were incubated with 0.2 μM ssDNA or dsDNA. Percent of the free DNA is calculated based on the gray scanning analysis. (G, H) Assembly of Cas protein complexes on gel filtration. The two peaks of Cas1/Cas2/Cas4/Spacer are evaluated by SDS-PAGE.