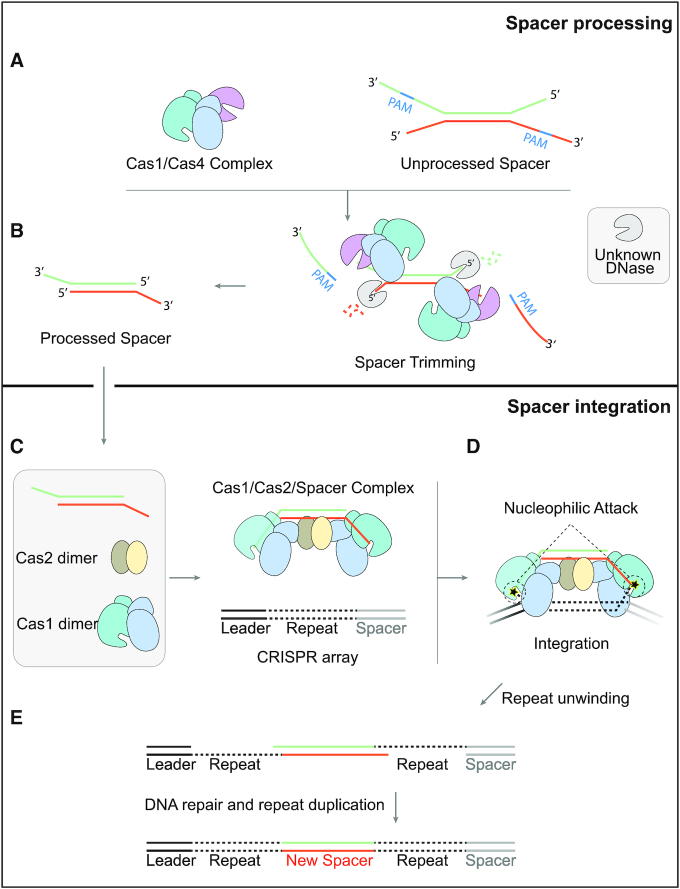
Figure 7.
A two-step sequential assembly model for the type I-D adaptation module of Synechocystis. (A) Synechocystis Cas4 and Cas1 easily assemble into a stable complex with a stoichiometry of 1 Cas4 to 2 Cas1 to recognize the unprocessed prespacer with a PAM sequence. (B) Cas4 trims the PAM-containing 3′ overhang of the prespacer in a PAM dependent manner, while an unknown DNase is thought to process the 5′ overhang. (C) Though Cas1 and Cas2 fail to form stable complex, Cas1, Cas2 and the processed prespacer are able to form a very stable ternary complex for integration. (D) The incoming prespacer is integrated into the CRISPR locus by nucleophilic attack of the two 3′-OH groups. (E) The gap-filling replication are performed by the host DNA replication machinery.