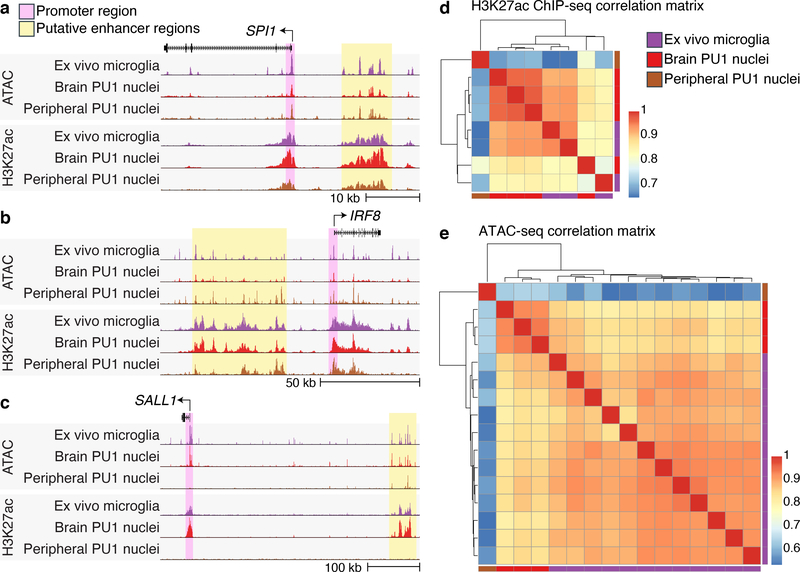
Fig. 3 |. Enhancer atlases from ex vivo microglia and PU.1 nuclei.
(a - c) Representative UCSC browser view of ATAC-seq and H3K27ac ChIP-seq from ex vivo microglia28, brain-derived PU.1 nuclei1 and peripheral-derived PU.1 nuclei at the (a) SPI1 locus, (b) IRF8 locus and the (c) SALL1 locus. (d) Pearson’s correlation of H3K27ac ChIP-seq signal from ex vivo microglia28 (N = 3), brain-derived PU.1 nuclei1 (N = 4) and peripheral-derived PU.1 nuclei (N = 1). (e) Pearson’s correlation of ATAC-seq signal from ex vivo microglia28 (N = 12), brain-derived PU.1 nuclei1 (N = 3) and peripheral-derived PU.1 nuclei (N = 1).