Figure 4. (A) Left: Neutrophil-Produced Siderocalin (Lcn-2, in Complex with Enterobactin, Ent; PDB: 3K3L) and Bacterial Response Exemplified with the Production of Ent and Salmochelin (Sal), Which Is Not Recognized by Lcn-2.
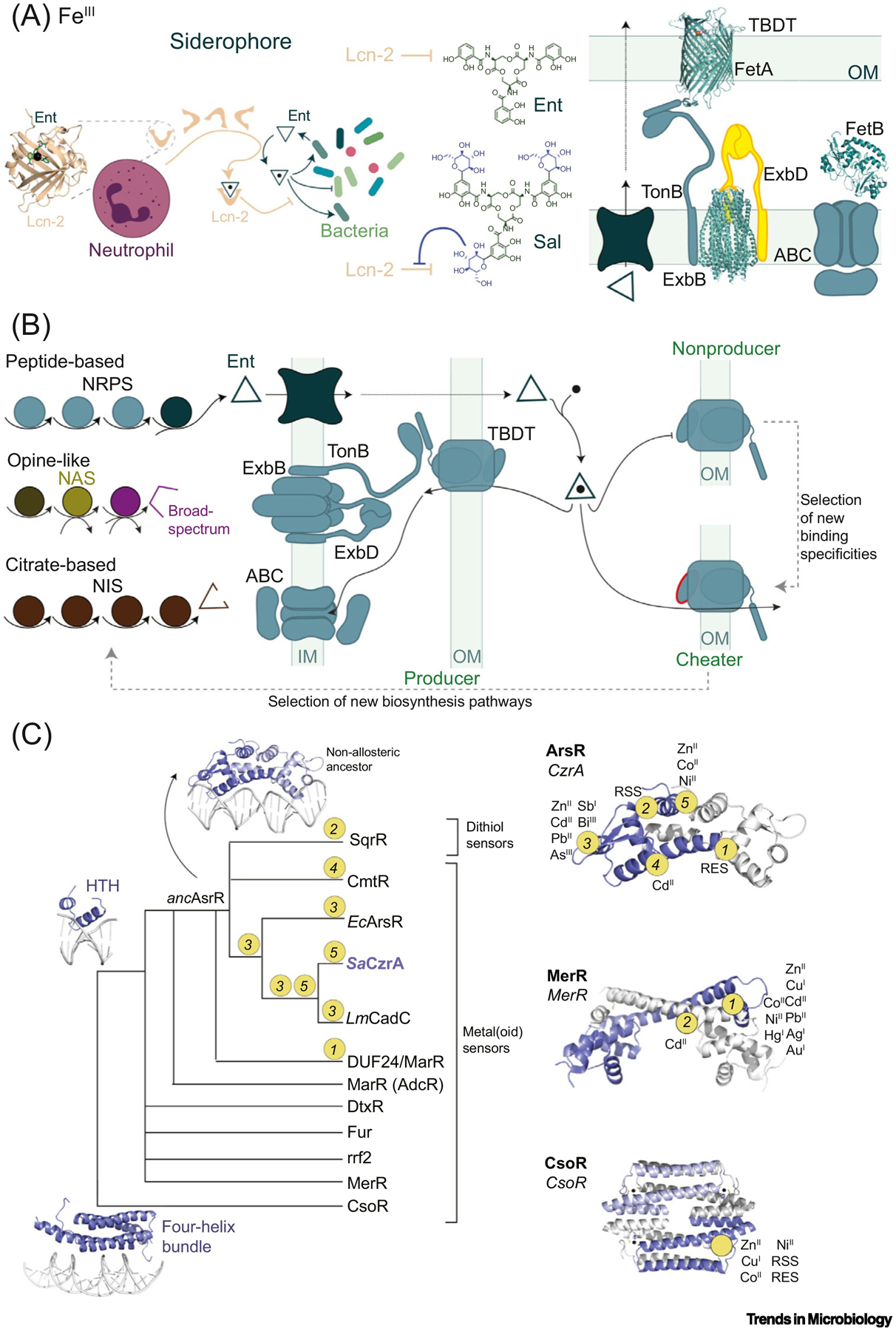
Right: Gram-negative bacterial import system exemplified by the TonB-dependent transporter (TBDT) ZnuD (PDB: 4RVW), ExbB/ExbD complex (PDB: 5SV1), and an ATP-binding cassette (ABC) transporter (showing the structure of solute-binding protein FepB (PDB: 2M6L). (B) Siderophore or metallophore production (left) and cellular uptake (right). In Gram-negative bacteria, siderophores are imported by TBDTs into the periplasm and then into the cytoplasm by ABC transporters. In non-siderophore-producing bacteria (cheaters), new binding affinities and specificities in TBDTs are selected for, allowing for adaptation to new niches [87]. This, in turn, exerts a selective pressure on siderophore-producing bacteria, resulting in new biosynthesis pathways that expand siderophore diversity. (C) Schematic representation of the evolution of transcriptional regulator families, as suggested by some studies [99,100,106,129]. Apart from the CsoR superfamily (which may have arisen from a four-helix bundle ancestor), most metallosensors share the helix-turn-helix (HTH) topology in a common ancestor. In the arsenic repressor (ArsR) superfamily, the five sensory sites evolved from a nonallosteric ancestor with distinct metal or reactive species affinities and reactivities, respectively. The approximate positions of inducer recognition sites and sensing specificities are shown in representatives of the ArsR (2M30), MerR (5CRL), and CsoR (4M1P) superfamilies [103,130]. Abbreviations: NIS, NRPS-independent siderophore; NRPS, non-ribosomal peptide synthesis; OM, outer membrane; RSS, reactive sulfur species; RES, reactive electrophile species.
