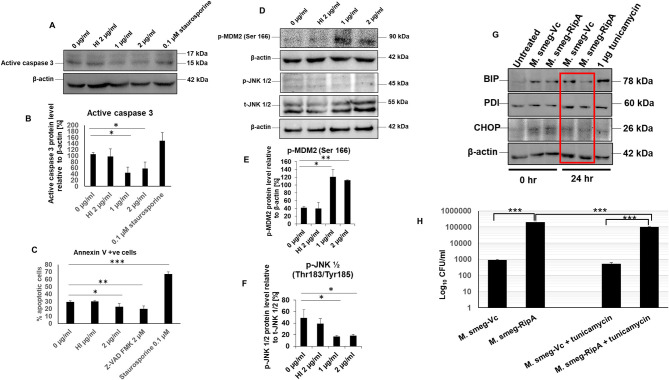
Figure 9.
RipA inhibits caspase-dependent apoptosis of RAW264.7 cells. (A) Western blots showing the level of active caspase 3 in RipA treated samples. Untreated and HI treated cells were used as negative controls. (B) Densitometric quantitation of active caspase 3 in RipA treated samples. (C) Flow cytometric analysis of early apoptotic cells in RipA treated macrophages. HI-treated cells were used as a negative control, whereas staurosporine and ZVAD-FMK served as controls for caspase-dependent apoptosis induction and repression. (D) Western blots showing the level of activated phospho MDM2 and inhibited phospho JNK 1/2, and total JNK 1/2. Total JNK 1/2 and β-actin were used as controls. (E,F) Densitometric quantification of pMDM2 and pJNK 1/2 protein bands are shown as bar graphs. (G). Western blots showing the levels of ER stress markers BIP, PDI, and CHOP. (H) RAW264.7 macrophage cells (2 × 106 cells) were infected with recombinant M. smegmatis expressing M. tb RipA and vector alone at MOI-1:10. Colony-forming units were determined after 24 h of infection. Recombinant M. smegmatis containing RipA showed higher survival inside macrophages in untreated, as well as tunicamycin treated cells demonstrating higher infectivity and virulence capacity of recombinant M. smegmatis containing RipA. Data are representative of 3 independent experiments and expressed as means±SD. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. controls.