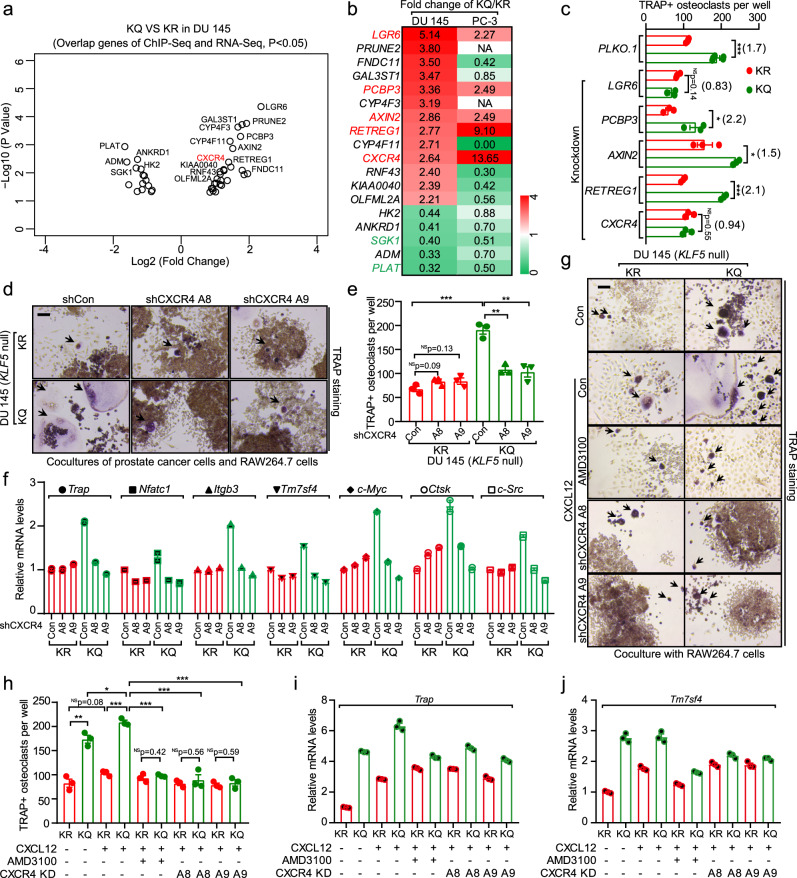
Fig. 5. Identification of CXCR4 as a functional effector of acetylated KLF5 in the induction of osteoclast differentiation.
a Differentially expressed genes between KLF5KQ- expressing (KQ) and KLF5KR-expressing (KR) DU 145 cells, as identified by ChIP-Seq and RNA-Seq analyses and illustrated by a volcano diagram according to their p-values and fold changes in RNA-Seq. Circles indicate genes whose promoter regions (−2500 to +500) had binding peaks in the ChIP-Seq assay. Details are available in Supplementary data 7. b Heatmap of genes in panel a with fold changes between KQ and KR in both DU 145 and PC-3 cells, as revealed by RNA-Seq analysis. Each rectangle is colored and the intensity is defined by the fold change of the gene expression levels in KQ versus KR cells. Red and green indicate upregulation and downregulation, respectively. Gene names in red and green indicate genes that are upregulated and downregulated, respectively, by KQ in both cell lines. c Functional screening for genes that mediate osteoclast differentiation induced by KLF5KQ (KQ)-expressing cells. Each of the KQ-upregulated genes from panel b was knocked down by a mixture of 2 or 3 shRNAs, each of which was confirmed to efficiently knock down its target gene, in both KQ and KR cells, and then co-cultured with RAW264.7 pre-osteoclasts. TRAP was stained to measure osteoclast differentiation. Numbers in parentheses indicate fold changes of TRAP + osteoclast numbers between KQ and KR. n = 5 wells for control shRNA PLKO.1 and n = 3 wells for other shRNAs. d, e TRAP staining (d) and statistical analyses of TRAP + osteoclasts (e) in co-cultures of KR- or KQ-expressing DU 145 cells with RAW264.7 pre-osteoclasts in the presence of 20 ng/ml Rankl for 6 days. Black arrows indicate TRAP + multinucleated osteoclasts. A8 and A9 are two different CXCR4 shRNAs. n = 3 wells per group in e. f Knockdown of CXCR4 selectively suppresses the expression of markers for osteoclast differentiation in KQ cells, as detected by real-time qPCR. g, h TRAP staining (g) and statistical analyses of TRAP + osteoclasts (h) in co-cultures of RAW264.7 and KR- or KQ-expressing DU 145 cells with inhibition of CXCR4 by either the AMD3100 inhibitor (500 ng/ml) or shRNAs (A8 and A9). The CXCR4 agonist ligand CXCL12 (100 ng/ml) was added as indicated. n = 3 wells per group in h. i, j Expression of osteoclast differentiation markers Trap (i) and Tm7sf4 (j) was detected by real-time qPCR in co-cultures from panel g. Scale bar, 100 μm. In panels c, e and h, data are shown in mean ± S.E.M. NS not significant; *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed Student’s t test). Source data are provided as a Source Data file.