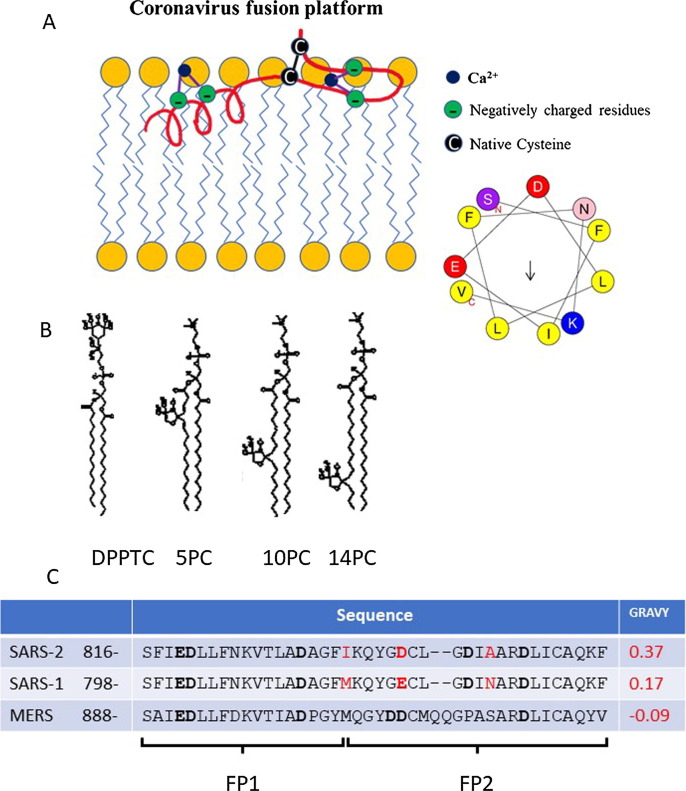
Figure 1.
(A). A model of the coronavirus FP or “platform” interacting with a lipid bilayer (adopted from our previous model18 using the structural information from CD spectroscopy in this study, and the molecular dynamics model.22 A Heliquest Plot23 for the first 11 amino acids is shown. (B) The structure of spin labeled lipids used: DPPTC, 5PC, 10PC and 14PC. (C) The sequence alignment of SARS-2, SARS-1, and MERS FPs, and their grand average of hydropathy (GRAVY) values (calculated using GRAVY Calculator, http://www.gravy-calculator.de/index.php). The residues in red highlight the difference between SARS-2 and SARS FP. The residues in bold are the negatively charged residues which are potential Ca2+ binding sites.