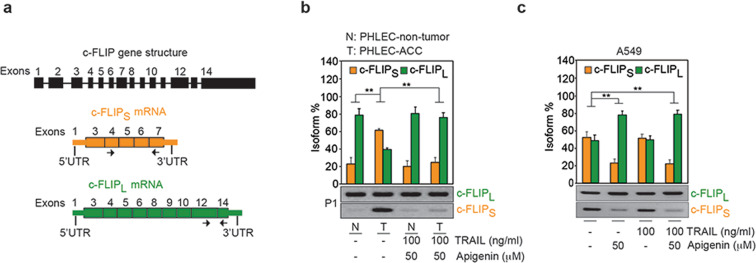
Fig. 4. Apigenin modulates alternative splicing of c-FLIP isoforms.
a Schematics of the c-FLIP gene structure including introns represented by a line and exons represented by filled numbered boxes. Splice isoforms of c-FLIP mRNA, c-FLIPS (in orange), and c-FLIPL (in green) including exons and UTRs represented by thinner filled boxes. b, c The alternative splicing of c-FLIPS and c-FLIPL was determined by RT-PCR using isoform-specific primers (indicated by the black arrows in [a]). Quantification of the isoforms observed in the gels was done by densitometry. The percent of each isoform was calculated as follows: (density of one isoform)/(density of the sum of the two isoforms) × 100. Bar plots summarize the quantitation and represent the mean ± SEM. The picture in the bottom is a representative image of an agarose gel depicting the c-FLIPS and c-FLIPL isoforms obtained by RT-PCR and separated by gel electrophoresis. b PHLECs isolated from paired nontumor (N) and adenocarcinoma tumor (T) biopsies from human patients (P1–P3) pretreated for 18 h with 50 μM apigenin or DMSO (-) followed by treatment with 100 ng/ml TRAIL for 6 h or controls treated only with DMSO (-) then, the alternative splicing of c-FLIPS and c-FLIPL was determined by RT-PCR using isoform-specific primers. c A549 NSCLC cells pretreated with 50 μM apigenin or DMSO (-) for 18 h followed by treatment with 100 ng/ml TRAIL for 6 h, apigenin or TRAIL alone; then, the alternative splicing of c-FLIPS and c-FLIPL was determined by RT-PCR using isoform-specific primers (black arrows shown in a). All data are expressed as the mean ± SEM. (Fig. 4b, n = 3; Fig. 4c, n = 4; **P < 0.01; and analyses were performed by two-way ANOVA followed by Tukey’s post hoc analysis).