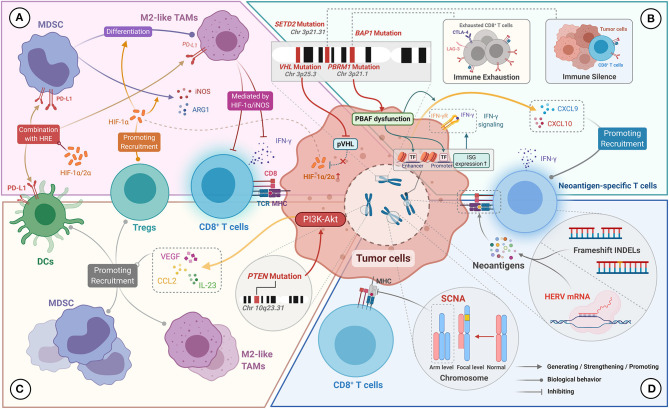
Figure 3.
Genomic characteristics of ccRCC for manipulating the TIME. (A) VHL mutations result in reduced pVHL production and reduced HIF-1α/2α degradation. Excess HIF-1α/2α upregulates the expression of PD-L1 on MDSC, M2-like TAMs, and DCs by binding to the hypoxia response element (HRE). HIF-1α promotes MDSCs to produce iNOS and Arg-1 and to differentiate into M2-like TAMs. HIF-1α promotes the recruitment of Tregs, and mediates the inhibitory effect of M2-like TAMs on the effector function of CD8+ TILs. (B) PBRM1 mutation leads to PBAF complex dysfunction, which upregulates interferon-stimulated gene (ISG) expression, thereby enhancing the tumor killing effect mediated by IFN-γ signaling. PBAF inactivation also promotes the secretion of CXCL9 and CXCL10 by tumor cells. BAP1 mutations are associated with the immuno-exhausted tumor microenvironment. SETD2 mutations are associated with the immune-silenced tumor microenvironment. (C) PTEN mutations activate the P13K-Akt pathway, resulting in upregulation of VEGF, CCL2, and IL-23 expression, which contribute to the recruitment of Tregs, DCs, and MDSCs. (D) Frameshift INDELs and HERV expression can generate abundant neoantigens, which stimulate the production of neoantigen-specific T cells. Arm level SCNA can disrupt the antigen-presenting capacity of MHC on tumor cells, resulting in inactivation of tumor-specific immune responses.