Abstract
SDS is widely used in sample preparation for proteomic research. However, SDS is incompatible with LC and electrospray ionization. SDS depletion is therefore required ahead of LC–MS analysis. Most of current SDS removal strategies are time consuming, laborious, and have low reproducibility. Here, we describe a method, SDS–cyclodextrin (CD)-assisted sample preparation, by which CD can bind to SDS and form CD–SDS complexes in solutions, allowing for direct tryptic digestion. We demonstrate that SDS–CD-assisted sample preparation is a simple, fast, and robust SDS-based sample preparation method for proteomics application.
Keywords: SDS, SDC, sample preparation
Abbreviations: ACN, acetonitrile; BAEE, Nα-benzoyl-l-arginine ethyl ester hydrochloride; CAA, 2-chloroacetamide; CD, cyclodextrin; DDA, data-dependent acquisition; FA, formic acid; FASP, filter-aided sample preparation; FDR, false discovery rate; FFPE, formalin-fixed and paraffin-embedded; iRT, independent retention time; iST, in-StageTip; PASEF, parallel accumulation—serial fragmentation; SCASP, SDS–CD-assisted sample preparation; SDB-RPS, poly(styrenedivinylbenzene)-reversed phase sulfonate; SDC, sodium deoxycholate; SP3, single-pot and solid-phase–enhanced sample preparation; SWATH-MS, Sequential Window Acquisition of All Theoretical Mass Spectra; TCEP, Tris(2-carboxyethyl)phosphine hydrochloride; TFA, trifluoroacetic acid
Graphical Abstract
Highlights
-
•
SCASP can effectively process SDS-containing samples.
-
•
SCASP is robust and easy to implement.
-
•
Superior performance of SCASP compared with current SDS-based methods.
-
•
SCASP is suitable for preparation of challenging samples.
In Brief
Cyclodextrins can be used for removal of SDS in protein solution, facilitating direct tryptic digestion. The SDS–cyclodextrin-assisted sample preparation method is a simple, robust, and reproducible strategy for proteomics research and highly suitable for challenging samples.
Sample preparation is a crucial step for LC–MS analysis, which largely determines the quality of the MS data. SDS is an effective surfactant to solubilize and extract proteins from various biological sources, which has considerable utility in both “bottom–up” and “top–down” proteomics (1). However, SDS is detrimental to chromatographic separation and causes severe suppression in electrospray mass spectrometry (MS) (2, 3). Besides, trypsin activity is inhibited even in low levels of SDS. Considerable efforts have been made to develop the strategies for SDS removal, including in-gel digestion (4), chromatography-based strategy (5), protein precipitation in organic solvents (6) or detergent precipitation with potassium chloride (7), filter-aided sample preparation (FASP) (8, 9), detergent removal spin column, and protein-binding microsphere beads–based strategies (10, 11, 12). However, each of these SDS removal methods has their drawbacks, such as labor or time intensive, sample loss, high cost, or low throughput. Cyclodextrins (CDs) are a family of cyclic oligosaccharides with a hydrophilic outer surface and a hydrophobic central cavity, which allows the formation of host–guest inclusion complexes with various compounds (13). This property of CDs makes it have a wide variety of applications in pharmaceuticals, drug delivery systems, cosmetics, and the food and chemical industries (14). SDS bears a hydrophilic head and a hydrophobic tail, which can be included in the internal cavity of CDs. Several studies showed that SDS formed complexes with CDs (15, 16). CDs were used for removal of SDS in peptide samples ahead of MS analysis (16). In this study, we showed that SDS in protein solutions can be “trapped” by CDs, and CD–SDS complexes can be removed by desalting. We employed this SDS removal strategy for in-solution tryptic digestion sample preparation, which we referred to as SDS–cyclodextrin-assisted sample preparation (SCASP). SCASP is extremely simple, robust, and highly reproducible and can be applied for samples from a variety of biological sources.
Experimental Procedures
Cell Culture
Human embryonic kidney 293T, human HeLa, human malignant melanoma A375, murine fibroblast L929, murine microglial BV2, and mouse neuronal HT-22 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Human T lymphocyte Jurkat and human B lymphocyte Raji cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were tested for Mycoplasma contamination. The cells were collected by centrifugation at 200g for 10 min, followed by three times wash using PBS. Cell viability and number counts were performed according to the manufacturer using a Countess automated cell counter (Life Technologies).
Murine L929 Cell Lysate Preparation
L929 cells were lysed with 1% sodium deoxycholate (SDC; V900388; Sigma)/10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; C4706; Sigma)/40 mM 2-chloroacetamide (CAA; 22790; Sigma)/100 mM Tris–HCl (pH 8.5). The lysates were kept at 37 °C for 30 min for reduction of disulfide bonds and alkylation of free cysteines and then sonicated to denature proteins and shear DNA. The protein concentrations determined by bicinchoninic acid protein assay kit–reducing agent compatible (23250; Thermo Fisher Scientific). The final protein concentration was adjusted to 5 μg/μl and stored at −80 °C.
Preparation of CD
α-CD, 2-hydroxypropyl-β-CD (HP-β-CD), and γ-CD were purchased from Sigma (α-CD, C4642; HP-β-CD, H107; and γ-CD, C4892). CDs were prepared to 250 mM in water and stored at 4 °C. α-CD and γ-CD is heated at 70 °C for 30 min for complete dissolution. We also tested α-CD (A600348), HP-β-CD (A600388), and γ-CD (A600350) from Sangon Biotech, and the reagents work as well as those from Sigma but at significantly lower costs.
Trypsin Activity Assay
Trypsin activity was measured by monitoring of the hydrolytic rate of Nα-benzoyl-l-arginine ethyl ester hydrochloride (BAEE) (B4500-10G; Sigma) into a UV-active product Nα-benzoyl-l-arginine using a UV spectrophotometer (Varian Eclipase) at 253 nm. Briefly, BAEE in 1% SDS was mixed with HP-β-CD at different CD:SDS mole ratios of 0, 0.72, 1.44, and 2.16 at room temperature. Trypsin was added, and the absorbance values at 253 nm were recorded. The first 3 min of reaction was used to calculate the slope of absorbance change at 253 nm versus time, which defines the trypsin activity.
L929 Lysate Digestion
About 10 μg of L929 lysate proteins (sample volume is 2 μl) were dissolved in 50 μl 1% SDC/100 mM Tris–HCl (pH 8.5) or 1% SDS/100 mM Tris–HCl (pH 8.5) and boiled for 10 min. Each experiment was performed in biological triplicates. SDC concentration was diluted to 0.5% with 50 μl HPLC water. The different volumes of 250 mM CD were added into the SDS samples, followed by mixing thoroughly. Trypsin (T6567; Sigma) was added at the ratio of 100:1 (micrograms of protein:micrograms of trypsin). Digestion was performed at 37 °C overnight unless indicated. After desalting, the 1 μg peptides were analyzed by TripleTOF 5600 (Sciex).
Acetone Precipitation and In-Solution Digestion
About 10 μg of L929 lysate proteins were dissolved in 50 μl 1% SDS/100 mM Tris–HCl (pH 8.5), and fourfold volumes of ice-cold 100% acetone were added. Each experiment was performed in biological triplicates. The samples were placed at −20 °C overnight. The pellets were collected by centrifugation at 12,000 rpm for 10 min at 4 °C, and supernatants were removed, and 50 μl solution was left while ensuring the pellets were not disturbed. The pellets were washed with 1 ml ice-cold 100% acetone for 3 times. The pellets were air dried at room temperature. The pellets were dissolved in 50 μl 1% SDC/100 mM Tris–HCl (pH 8.5), followed by vertexing for 30 min at 37 °C. SDC concentration was diluted to 0.5% with 50 μl HPLC water, followed by addition of trypsin. Digestion was performed at 37 °C overnight. After desalting, the 1 μg peptides were analyzed by TripleTOF 5600.
FASP
FASP was performed as previously described (9). Briefly, 10 μg of L929 lysate proteins were dissolved in 100 μl 1% SDS/100 mM Tris–HCl (pH 8.5) and then loaded into the Amicon Ultra-0.5 ml filter (30 K; Millipore). The samples were concentrated by centrifugation at 12,000 rpm for 10 min, and then 8 M urea was added into the filter. After centrifugation for 10 min, 8 M urea was added. The addition of 8 M urea was repeated for 3 times, and subsequently, 100 mM Tris–HCl (pH 8.5) was added into the filter. Trypsin was added at the protein:trypsin ratio of 100:1. Digestion was performed at 37 °C overnight. After desalting, the 1 μg peptides were analyzed by TripleTOF 5600.
Single-Pot and Solid-Phase–Enhanced Sample Preparation
About 5 μg of L929 lysate proteins were dissolved in 50 μl 1% SDS/100 mM Tris–HCl (pH 8.5) and then followed by the single-pot and solid-phase–enhanced sample preparation (SP3) method (17). Sera-Mag SpeedBeads (GE Healthcare; catalog no. 45152105050250) and Sera-Mag SpeedBeads (GE Healthcare; catalog no. 65152105050250) were mixed in equal amounts. About 500 μg of prepared SP3 beads were added into protein solutions and pipette mix to homogenize the solution. About 50% of ethanol (final concentration) was added into the protein solution containing the SP3 beads. The mixture was shaken at 1000 rpm at 24 °C for 5 min. The supernatant was removed, and 80% ethanol was added to rinse the beads. The beads were washed with 80% ethanol for two more times, and 100 μl of 100 mM Tris–HCl (pH 8.5) containing trypsin was added to the beads. Digestion was performed at 37 °C overnight. The peptides were cleaned up using poly(styrenedivinylbenzene)-reversed phase sulfonate (SDB-RPS) StageTips.
Cell Digestion
Cells were washed with ice-cold PBS for 3 times and collected by centrifugation for 5 min at 1000g. Cell numbers were counted, and 5 × 104 cells were dissolved in 50 μl 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5), 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) and 8 M urea/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers, respectively. Each experiment was performed in biological triplicates. The SDC and SDS samples were boiled for 10 min, whereas the urea samples were kept at 37 °C for 30 min. SDC concentration was diluted to 0.5% with 50 μl HPLC water, whereas 12.5 μl 250 mM HP-β-CD were added into the SDS samples and pipette mix to homogenize the solution. Urea concentration was diluted to below 2 M with 100 mM Tris–HCl (pH 8.5). Trypsin was added at the protein:trypsin ratio of 100:1 (assuming 200 pg for 1 cell; BioNumber 109385). Digestion was performed at 37 °C overnight. After peptide cleanup, the 1 μg peptides were analyzed by TripleTOF 5600.
The Whole-Proteome Analysis
About 5 × 105 HeLa cells were dissolved in 100 μl 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers. Sonication of lysate can significantly increase identified peptides before heat (supplemental Fig. S9). The protein mixtures were heated at 95 °C for 10 min. Subsequently, SDC concentration was diluted to 0.5% with 50 μl HPLC water, whereas 30 μl 250 mM HP-β-CD were added into the SDS samples and pipette mix to homogenize the solution. FASP was performed as described previously. Trypsin was added at the protein:trypsin ratio of 100:1. Digestion was performed at 37 °C overnight. The peptides were cleaned up using SDB-RPS StageTips before high-pH reverse phase chromatography fractionation. About 20 fractions were generated, and each fraction was considered to contain 5 μg peptides. About 1 μg of peptides were analyzed on TripleTOF 5600.
Yeast
The budding yeast Saccharomyces cerevisiae strain was grown in the yeast extract peptone dextrose media, incubated (30 °C, 230 rpm) and harvested at an absorbance of ≈1.0 at 600 nm. Cell numbers were counted and washed 3 times with cold water followed by centrifugation (4000g, 5 min, and 4 °C). The pellets were stored at −80 °C until used. About 107 yeast cells were dissolved in 100 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 100 μl 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5), and boiled for 10 min. Each experiment was performed in biological triplicates. The SDC and SDS samples were boiled for 10 min. After centrifugation, the supernatants were transferred to the new tubes. SDC concentration was diluted to 0.5% with 50 μl HPLC water, whereas 25 μl 250 mM HP-β-CD were added into the SDS samples and pipette mix to homogenize the solution. Trypsin was subsequently added at protein:trypsin ratio of 100:1 (assuming 5 pg protein per yeast cell, BioNumber 110550). Digestion was performed at 37 °C overnight. After peptide cleanup, the 2 μg peptides were analyzed by TripleTOF 5600.
Plant Leaf
Rice Arabidopsis thaliana leaves were abundantly washed with water, blot dried with filter paper, frozen in liquid nitrogen, and stored at −80 °C until extraction. Leaf tissue (2 g fresh weight) was crushed in a precooled mortar with liquid nitrogen until a fine powder was formed. About 1 mg powder was dissolved with 100 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) followed by boiling for 10 min. Each experiment was performed in biological triplicates. After centrifugation, the supernatants were transferred to the new tubes. SDC concentration was diluted to 0.5% with 50 μl HPLC water, whereas 25 μl 250 mM HP-β-CD were added into the SDS samples. Trypsin was subsequently added at protein:trypsin ratio of 100:1 (assuming 10 μg proteins was extracted from 1 mg leaf powder). Digestion was performed at 37 °C overnight. After peptide cleanup, the 2 μg peptides were analyzed by TripleTOF 5600.
Murine Tissues Digestion
The C57BL/6 mice of postnatal 50 days were used for tissue dissection. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee at Xiamen University. Tissues were snap frozen in liquid nitrogen upon dissection. About 10 mg of tissue was used for SCASP and the SDC method. Taking into account that protein extraction efficiency from tissues is about 1%, 100 μg proteins can be extracted from 10 mg of tissue. About 500 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) were added into the tissue samples, followed by homogenization on the Scientz-48 High Throughput Tissuelyser (Scientzbio). Around 10 μg proteins were used for digestion, and the volumes of reactions were adjusted to 100 μl by adding1% SDS or SDC. Each experiment was performed in biological triplicates. The SDC and SDS samples were boiled for 10 min. SDC concentration was diluted to 0.5% with 50 μl HPLC water, whereas 25 μl 250 mM HP-β-CD were added into the SDS samples. Trypsin was subsequently added at protein:trypsin ratio of 100:1. Digestion was performed at 37 °C overnight. After peptide cleanup, the 2 μg peptides were analyzed by TripleTOF 5600.
Formalin-Fixed and Paraffin-Embedded Murine Tissues
For laser-capture microdissection, formalin-fixed and paraffin-embedded (FFPE) tissues were mounted on polyethylene naphthalate membrane slides to enable efficient cutting and collection. An area of approximately 5 × 5 mm (10 μm thick section) was collected into 1 ml Eppendorf tubes. Assuming the average mammalian cell volume is 2250 μm3 (BioNumber ID: 100434), there are about 105 cells in one section. Xylene-mediated deparaffinization was accomplished by a 1 ml xylene incubation at 37 °C for 1 h. After vertexing and 5 min centrifugation at 20,000g, xylene was removed. A second incubation with 1 ml xylene was conducted at 37 °C for 30 min, followed by centrifugation at 20,000g for 5 min. After xylene was removed, 1 ml of 100% ethanol was added for 5 min at room temperature. After steps of vortexing and centrifugation like mentioned previously, ethanol was removed. Ethanol wash repeated one more time. Ethanol was discarded, and the remaining pellets were dried for 30 min at room temperature.
About 100 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) were added into one section for protein extraction, followed by 95 °C for 30 min, and 1% SDC was diluted into 0.5% SDC with water, followed by the addition of trypsin at protein:trypsin ratio of 100:1 (assuming 200 pg for one cell). About 25 μl 250 mM CD was added into 1% SDS, and trypsin was subsequently added at protein:trypsin ratio of 100:1 (assuming 200 pg for one cell). Digestion was performed at 37 °C overnight. After peptide cleanup, the peptides corresponding to 10,000 cells were analyzed by TripleTOF 5600, and the peptides corresponding to 1000 cells were analyzed by timsTOF Pro.
Vesicular Stomatitis Virus Infection
About 2 × 107 human monocyte THP-1 cells were infected with one multiplicities of infection vesicular stomatitis virus (VSV) for 0, 2, 4, 6, and 8 h in biological duplicates. After treatment, THP-1 cells were harvested by centrifugation and washed with PBS for 5 times. About 1 × 105 cells were dissolved in 100 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) and boiled for 10 min. After cooling to room temperature, 25 μl 250 mM CD was added into 1% SDS, and trypsin was subsequently added at protein:trypsin ratio of 100:1 (assuming 200 pg for one cell). Digestion was performed at 37 °C overnight. After peptide cleanup, the 200 ng peptides were analyzed by timsTOF Pro.
Enrichment of Membrane Proteins
Membrane fractions of cells were enriched as previously described (18). Briefly, 500 μl of ice-cold water were added to 106 cells, and the samples were pipetted up and down 10 times, incubated for 10 min on ice, and frozen for 1 min in liquid nitrogen. The frozen samples vortexed 1500 rpm for 5 min at room temperature. Pellets were collected by centrifugation for 10 min at 10,000g at 4 °C. The pellets were washed with 500 μl of water, incubated, frozen, and thawed as the step mentioned previously. The pellets were washed with water for 3 times. After 500 μl of water were added to the pellets, the sample was pipetted up and down 20 times, incubated for 10 min on ice, centrifugated for 20 min at 100,000g at 4 °C. After three steps of water washes and super-high centrifugations, the pellets were subjected to SCASP.
Desalting
For all peptide samples, we used in-house made StageTips packed with SDB-RPS (2241, 3 M) material for desalting. About 1% trifluoroacetic acid (TFA; T6508; Sigma) was added into the reactions to stop digestion. For SDC-based digestion, centrifugation was performed before loading the peptides. For SCASP, no centrifugation was performed if there was no apparent pellet. The SDB-RPS StageTips were conditioned with 100 μl 100% acetonitrile (ACN) (3485; Sigma). The peptides were loaded into StageTips, followed by centrifugation at 4000g for 5 min. StageTips were washed twice with 100 μl 1% TFA/ isopropyl alcohol (I9030; Sigma), and then washed with 100 μl 0.2% TFA. Elution of peptides was performed using 80% ACN/5% ammonia water. All eluted materials were collected in glass vials (A3511040; CNW Technologies) and dried at 45 °C using a SpeedVac centrifuge (Eppendorf Concentrator Plus; 5305).
High-pH Reversed Phase Fractionation
High-pH fractionation was performed on a Dionex Ultimate 3000 system (Thermo Fisher Scientific). About 100 μg peptides were loaded and separated on a 2.1 × 100 mm C18 reversed-phase column (ACQUITY UPLC BEH) packed with 1.7 μm particles. A 60 min gradient was delivered as followed: 5 to 25% buffer B (buffer B: 10 mM ammonium formate, 40% ACN, 12.5% ammonia solution; buffer A: 20 mM ammonium formate, pH 10) in 20 min, then increased to 45% in 40 min and to 90% in 1 min. The resulting 60 fractions were pooled to 20 fractions. The pooling procedure was performed as follows: fraction x was pooled with fractions x + 10 and x + 20. The pooled fractions were desalted with SDB-RPS StageTips and evaporated using vacuum centrifugation.
MS
Data-Dependent Acquisition on TripleTOF 5600
Peptides were dissolved in 0.1% formic acid (FA) (06440; Sigma) and analyzed by MS. An Ultra 2D Plus (Eksigent) system was coupled to the TripleTOF 5600 through a Nanospray III source. Peptides first bound to a 5 × 300 μm trap column packed with Zorbax C18 5-μm 300-Å resin (5065-9913; Agilent) using 0.1% (v/v) FA/2% ACN in water at 10 μl/min for 5 min, and then separated using 30, 45, or 60 min gradient depending on the complexity of samples from 2 to 35% buffer B (buffer A: 0.1% [v/v] FA, 5% dimethyl sulfoxide in water; buffer B: 0.1% [v/v] FA, 5% dimethyl sulfoxide in ACN) on a 30 cm × 75 μm in-house pulled emitter-integrated column packed with Magic C18 AQ 3-μm 200-Å resin. The column temperature was kept at 50 °C by a column heater (PST_CHC-RC; Phoenix S&T) and a controller (PST-BPH-20; Phoenix S&T). MS1 spectra were collected in the range 350 to 1250 m/z for 250 ms, and up to 20 most intense precursors with charge state two to five were selected for fragmentation, and MS2 spectra were collected in the range 100 to 1800 m/z for 100 ms in high-sensitivity mode. Exclusion time for selection of precursor ions is 20 s.
Sequential Window Acquisition of All Theoretical Mass Spectra
Peptides were dissolved in 0.1% FA containing independent retention time (iRT) peptides and analyzed by Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH-MS). For SWATH-MS, an MS1 scan records a 350 to 1250 m/z range for 250 ms, and a 100 to 1800 m/z range was recorded for 33.3 ms in the high-sensitivity mode MS2 scan. One MS1 scan was followed by 100 MS2 scans, which covered a precursor m/z range from 400 to 1200. The variable windows of SWATH-MS were as follows: 399.5 to 409.9, 408.9 to 418.9, 417.9 to 427.4, 426.4 to 436, 435 to 443.6, 442.6 to 450.8, 449.8 to 458, 457 to 464.8, 463.8 to 471.1, 470.1 to 476.9, 475.9 to 482.8, 481.8 to 488.6, 487.6 to 494, 493 to 499, 498 to 504.4, 503.4 to 509.3, 508.3 to 514.3, 513.3 to 519.2, 518.2 to 524.2, 523.2 to 529.1, 528.1 to 534.1, 533.1 to 539, 538 to 543.5, 542.5 to 548.5, 547.5 to 553, 552 to 558, 557 to 562.5, 561.5 to 567, 566 to 571.5, 570.5 to 576, 575 to 580.5, 579.5 to 585, 584 to 589.5, 588.5 to 594, 593 to 598, 597 to 602.5, 601.5 to 607, 606 to 611.1, 610.1 to 615.6, 614.6 to 620.1, 619.1 to 624.6, 623.6 to 628.6, 627.6 to 633.1, 632.1 to 637.6, 636.6 to 642.1, 641.1 to 646.6, 645.6 to 651.1, 650.1 to 655.6, 654.6 to 660.1, 659.1 to 665.1, 664.1 to 669.6, 668.6 to 674.5, 673.5 to 679, 678 to 684, 683 to 688.5, 687.5 to 693.4, 692.4 to 698.4, 697.4 to 703.3, 702.3 to 708.7, 707.7 to 713.7, 712.7 to 719.1, 718.1 to 724.5, 723.5 to 729.9, 728.9 to 735.3, 734.3 to 740.7, 739.7 to 746.5, 745.5 to 751.9, 750.9 to 757.8, 756.8 to 763.6, 762.6 to 769.5, 768.5 to 775.3, 774.3 to 781.2, 780.2 to 787, 786 to 793.3, 792.3 to 800.1, 799.1 to 806.4, 805.4 to 813.1, 812.1 to 820.3, 819.3 to 827.5, 826.5 to 835.2, 834.2 to 843.3, 842.3 to 851.4, 850.4 to 859.9, 858.9 to 868.9, 867.9 to 878.4, 877.4 to 888.3, 887.3 to 899.1, 898.1 to 910.3, 909.3 to 922.9, 921.9 to 936, 935 to 949.5, 948.5 to 963.4, 962.4 to 978.7, 977.7 to 994.9, 993.9 to 1015.6, 1014.6 to 1042.2, 1041.2 to 1070.1, 1069.1 to 1100.7, 1099.7 to 1140.7, and 1139.7 to 1196.5.
timsTOF Pro
LC was performed on an ultrahigh pressure nanoflow chromatography system (Elute UHPLC; Bruker Daltonics). Peptides were separated on a reversed-phase column (40 cm × 75 μm i.d.) at 50 °C packed with Magic C18 AQ 2.5-μm 200-Å resin with a pulled emitter tip. A solution is 0.1% FA in water, and B solution is 0.1% FA in ACN. The gradient time is 60 min, and the total run time is 75 min including washes and equilibration. Peptides were separated with a linear gradient from 0 to 5% B within 5 min, followed by an increase to 30% B within 55 min and further to 35% B within 5 min, followed by a washing step at 95% B and re-equilibration. LC was coupled online to a hybrid trapped ion-mobility spectrometry quadrupole time-of-flight mass spectrometer (Bruker timsTOF Pro) via a CaptiveSpray nanoelectrospray ion source. We performed data-dependent acquisition (DDA) in parallel accumulation—serial fragmentation (PASEF) mode with ten PASEF scans per topN acquisition cycle. Singly charged precursors were excluded by their position in the m/z-ion mobility plane and precursors that reached a “target value” of 20,000 au were dynamically excluded for 0.4 min. For data-independent acquisition, we adopted the isolation scheme of 25 Da × 32 windows to cover 400 to 1200 mz.
Bioinformatics
Processing of DDA Data
DDA wiff files from TripleTOF 5600 and ddaPASEF.d files from timsTOF Pro were converted to centroid mzML files using MSConvert (version 3.0.19311). mzML files were searched using MSFragger (version 2.3) against an UniprotKB/Swiss-Prot murine or human or yeast protein database (murine and human databases were downloaded in June 27, 2019, and yeast and rice databases were downloaded in April 17, 2020) appendant with common contaminants and reversed sequence decoys (40,963 entries including decoys for human; 34,269 for mouse; 12,330 for yeast; 75,000 for rice) through the FragPipe interface (https://fragpipe.nesvilab.org/). The search parameters were set as followed: precursor monoisotopic mass tolerance “50 ppm” for 5600 and “15 ppm” for timsTOF, fragment mass tolerance “0.1 Da” for 5600 and “0.05 Da” for timsTOF, modification “57.021464@C,” potential modification mass “15.994915@M,” cleavage “semi” and maximum missed cleavage sites “1”. PeptideProphet, ProteinProphet, and false discovery rate (FDR) filtering were performed by philosopher software (version 3.2.2) (https://github.com/Nesvilab/philosopher) (19) through the FragPipe interface (https://fragpipe.nesvilab.org/). The pep.xml search results were validated and scored using PeptideProphet followed by analysis with ProteinProphet. The precursor ions and proteins were filtered at 1% FDR.
Analysis of SWATH-MS Data
SWATH-MS wiff files were converted to centroid mzXML files using MSConvert (version 3.0.19311), which were subjected to DIA-Umpire software (version 2.1.6) analysis. Signal-extraction module of DIA-Umpire was used to generate pseudo-DDA mgf files. These mgf files were converted to mzML files, which are subjected to database search using MSFragger (version 2.3). Database search settings are the same as those used in DDA searches. The peptide ions that pass 1% protein FDR were built as a spectral library with SpectraST. The spectral library–based targeted analysis of SWATH-MS was performed using the QuantPipe (20) tool based on the OpenSWATH-PyProphet-Tric workflow (21, 22, 23). The results were filtered at 1% global protein FDR.
Building of the Spectral Library From ddaPASEF Data by Skyline
The combined interact.pep.xml files were input into Skyline (24) (version 20.1) to generate the spectral library. The cutoff values of PeptideProphet probability correspond to 1% protein FDR. The background proteome is the protein FASTA file filtered at 1% protein FDR generated by Philosopher software.
Targeted Analysis of diaPASEF Data by Skyline
The settings of Skyline software are described later. The “Peptide Settings” are listed as follows: “Digestion”—“Trypsin(semi)[KR|P],” “Max missed cleavages—2,” “Background proteome is the protein FASTA file filtered at 1% protein FDR generated by philosopher software.”—“Enforce peptide uniqueness by Proteins”, “Prediction”—”Retention time predictor”—“windows”—“10 min”, “iRT Calculator”—“Biognosys-11(iRT-C18)”, “use spectral library ion mobility values when presented”—checked, “Resolving power”—“50”, “Library”—“selected the prebuilt spectral library”. The “Transition Settings” are listed as follows: “Filter”—“Precursor charges 2, 3, 4, 5”; “Ion charges 1, 2”—“Ion types y, b”, “Product ion selection”—“From ion 1”—“to last ion”, “Library”—“Pick 6 product ions”, “Full-Scan”—“MS/MS filtering”—“Acquisition method DIA”—“Product mass analyzer TOF”—“Isolation scheme diaPASEF”—“Resolving power 30,000”—“Retention time filtering”—“Use only scans within 5 min”. The precursor ions in the spectral library were input into the target list though the spectral library explorer, and “decoy peptides” were then added into the target list. The THP-1 assay library contained 7015 proteins and 148,325 peptides, including decoys, 172,045 precursor ions, and 1027,123 transitions. Targeted analysis of diaPASEF data was accomplished by “Reintegrate”—“Peak scoring model”—“‘mProphet”. The identified precursor ions were exported by “File”—“Export”—“Report”. The resulting precursor ions were filtered at 0.01 of q value that is corresponding to 1% FDR. Protein intensities were calculated by summing the top three peptide intensities using InfernoRDN software (25) (https://github.com/PNNL-Comp-Mass-Spec/InfernoRDN).
Statistical Analysis and Plots
Protein intensities were input into Perseus (26) (version 1.6.10.43). Protein abundances were normalized with total intensities of all proteins per run and then log2 transformed. The Pearson correlation analysis, principal component analysis, hierarchical clustering, and differential expression analysis were performed with default settings.
Results
Establishment and Characterization of the SCASP Method
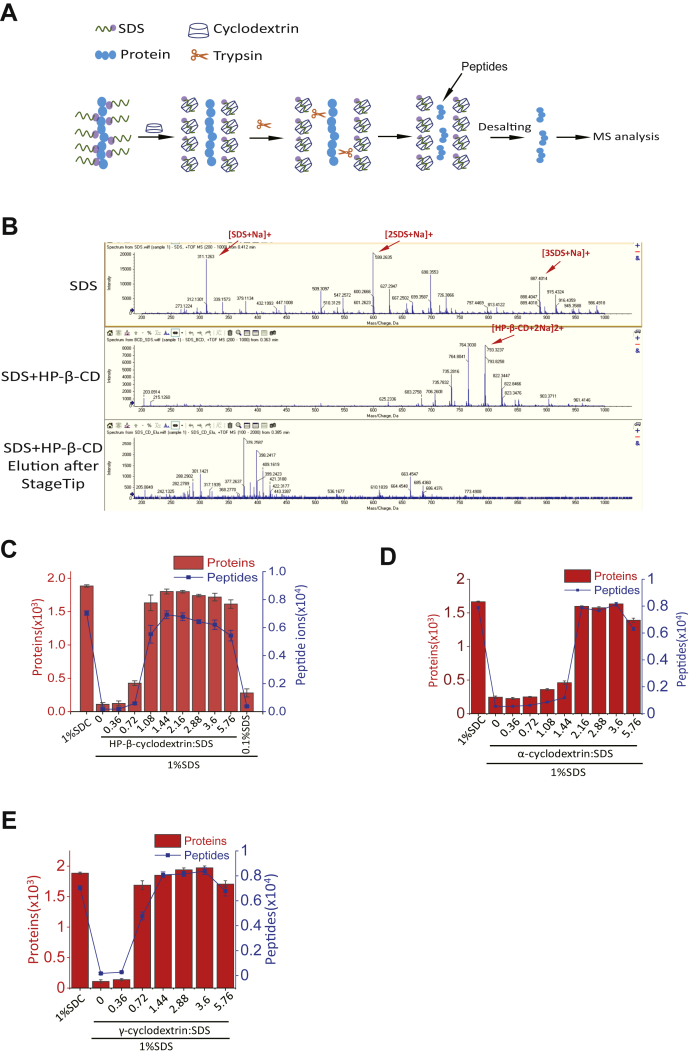
We hypothesize that SDS in solutions can be removed by addition of CDs, thereby facilitating direct tryptic digestion (Fig. 1A). To test this, we mixed SDS with HP-β-CD. SDS can be almost entirely removed by adding HP-β-CD, and extra HP-β-CD can be eliminated by desalting (Fig. 1B). Specifically, although the maximum peak intensity is about 2500 counts per second in the elution after StageTip cleanup, which is far lower than 20,000 and 8000 counts per second in SDS and SDS + HP-β-CD, there are no SDS or HP-β-CD peaks in the elution of StageTip. To determine the ratios of CDs to SDS that facilitate tryptic digestion, we added different amounts of HP-β-CD into 10 μg murine L929 cell lysate that is dissolved in 1% SDS buffer, followed by in-solution tryptic digestion. After peptide cleanup, the approximate 1 μg peptides were analyzed by DDA MS.
Fig. 1.
Establishment of the SCASP method.A, the principles of the SCASP method. B, MS spectra of SDS and inclusion complex of SDS and 2-hydroxypropyl-β-cyclodextrin (HP-β-CD). About 50 μl 1% SDS were mixed with or without 15 μl 250 mM HP-β-CD, followed by dried samples. The dried samples were dissolved in 0.1% FA and analyzed by direct infusion into TripleTOF 5600. SDS–HP-β-CD mixtures were desalted using SDB-RPS StageTips, and elutes were dried and dissolved in 0.1% FA. The elutes were analyzed by direct infusion into TripleTOF 5600. MS spectra of SDS and HP-β-CD were labeled in red arrows. C, numbers of identified proteins and peptide ions from the starting material of 10 μg L929 lysate proteins using the SDC-based method and the SDS-based method. In the SDS-based strategy, indicated different ratios of HP-β-CD:SDS were employed. After peptide cleanup, 1 μg peptide was analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. D, numbers of identified proteins and peptide ions from the starting material of 10 μg L929 lysate proteins using the SDC-based method and the SDS-based method. In the SDS-based method, indicated different ratios of HP-β-CD:SDS were employed. After peptide cleanup, 1 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. E, numbers of identified proteins and peptide ions from the starting material of 10 μg L929 lysate proteins using the SDC-based method and the SDS-based method. In the SDS-based method, indicated different ratios of γ-CD:SDS were employed. After peptide cleanup, 1 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. FA, formic acid; FDR, false discovery rate; SCASP, SDS–cyclodextrin (CD)-assisted sample preparation; SDB-RPS, poly(styrenedivinylbenzene)-reversed phase sulfonate StageTip; SDC, sodium deoxycholate.
The SDC-based strategy, which was also called as the in-StageTip (iST) method (27), identified 1884 ± 17 (average ± SD in biological triplicates) proteins and 7040 ± 160 peptide ions at 1% protein and precursor ion FDR (Fig. 1C and supplemental Table S1). In comparison, the SDS-based approach identified 109 ± 26 proteins and 173 ± 72 peptide ions. We diluted 1% SDS into 0.1% SDS, resulting in 280 ± 60 proteins and 377 ± 63 peptide ions. When the HP-β-CD was added, the number of identified proteins and peptide ions increased compared with the SDS-based digestion without CDs. The more HP-β-CD was added, the more proteins and precursors were identified. When the ratio of HP-β-CD to SDS is 1.44, the highest numbers of identified proteins and peptide ions, 1802 ± 117 and 6917 ± 277, were achieved (supplemental Table S1). As the ratio of HP-β-CD to SDS is 2.16, 1799 ± 20 proteins and 6778 ± 276 peptide ions were identified, which is a slight reduction compared with the ratio of 1.44. Identified peptide sequences also showed the similar pattern (supplemental Fig. S1 and supplemental Table S1). CD to SDS ratio above 2.16 leads to a decrease of both protein and precursor numbers. CD can interact with the amino acid of the proteins, so excess CD probably masks the cleavage site of trypsin (3). In line with the masking effect of CD, the percentage of missed cleavages of peptides increased from 9.46 to 11.87% when the ratio of HP-β-CD:SDS increased from 1.08 to 5.76 (supplemental Fig. S2). This masking effect probably hinders the trypsin cleavage, resulting in a decrease of both protein and peptide numbers. The average number of proteins identified by the SDS-HP-β-CD digestion strategy decreased by 4.3% (82 of 1884) and 5.5% (85 of 1884) at CD:SDS ratio of 1.44 and 2.16 compared with those using the SDC method, respectively. The average numbers of precursor decreased by 1.7% (123 of 7040) and 3.7% (262 of 7040) compared with those using the SDC method, respectively. These results suggested that the 1.5 to 2.0 times of the amount of HP-β-CD to SDS enables the complete CD binding of SDS and facilitate subsequent tryptic digestion, which yielded comparable protein and peptide identifications with the SDC-based method. We examined the effect of NaCl on the complex formation of CD and SDS. As shown in supplemental Fig. S3, up to 100 mM NaCl does not affect the formation of CD–SDS complexes, but higher than 100 mM NaCl will decrease the performance of SCASP. However, we cannot exclude the possibility that reduced performance of SCASP is attributed to impaired trypsin activity at the high salt concentration.
To determine the trypsin activity at different ratios of HP-β-CD to SDS, we took BAEE as the substrate to perform the enzyme assay. The results of the enzyme activity assay are shown in supplemental Fig. S4 and supplemental Table S1. Trypsin activity was nearly completely blocked at 1% SDS condition, whereas the addition of CD to the reactions restored its activity. At the CD:SDS ratio of 0.72, trypsin gained 60% of the original activity. When CD:SDS ratios increased to 1.44 and 2.16, trypsin showed 96% and 93% of original activity, respectively. These results were consistent with identified peptides and proteins under different CD to SDS ratios.
We also test the effects of α-CD and γ-CD in the SDS–CD-based digestion methods. Both α-CD and γ-CD can form the complexes with SDS in solutions, resulting in the environment where the trypsin functions normally (Fig. 1, D and E and supplemental Table S1). The optimal ratios of α-CD and γ-CD to SDS are 2.6 to 3.6 and 1.44 to 3.6, respectively. From α-CD to γ-CD, the amount of CDs required for SDS–CD complex formation gradually decreased, which is probably attributed to the varying cavity size of different CDs (supplemental Fig. S5).
We subsequently evaluated the reproducibility of the SCASP method. SCASP and SDC methods were used to digest 10 L929 lysate samples, respectively, and the tryptic peptides were analyzed by SWATH-MS. The peptide and protein intensities of 20 samples were compared (Fig. 2, A and B and supplemental Table S1). The Pearson correlations between ten samples using the SDC method are from 0.81 to 0.94 at peptide levels and from 0.92 to 0.98 at protein levels, whereas those values using SCASP are from 0.84 to 0.95 at peptide levels and 0.94 to 0.98 at protein levels. These results demonstrated that SCASP showed good reproducibility similar to the SDC method.
Fig. 2.
Characterization of SCASP.A, correlation analysis of protein intensities from ten samples using the SDC-based method and SDS-based method. About 10 μg L929 lysate proteins were dissolved in SDS or SDC buffers. After peptide cleanup, 1 μg peptides were analyzed by SWATH-MS. The identified precursor ions and proteins were filtered at 1% global protein FDR. B, correlation analysis of peptide intensities from ten samples using the SDC-based method and SDS-based method. C, numbers of identified peptide ions and proteins at different enzyme:substrate ratios using the SDC-based method and SCASP. About 10 μg L929 lysate proteins were dissolved in SDS or SDC buffers, and indicated amount of trypsin was added. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. D, numbers of identified peptide ions and proteins using different amount of starting materials. About 10, 50, and 100 μg L929 lysate proteins were dissolved in SDS or SDC buffers, followed by digestion. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. E, numbers of identified peptide ions and proteins using the SDC-based method and SCASP under the conditions of various incubation time and trypsin or trypsin plus lysine-C. About 10 μg L929 lysate proteins were dissolved in SDS or SDC buffers, and trypsin or trypsin plus lysine-C were added. The digestions were incubated for indicated varying time. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. F, numbers of identified peptide ions and proteins using indicated different SDS concentrations. About 10 μg L929 lysate proteins were dissolved in 1%, 5%, and 10% SDS buffers, and HP-β-CD was added to keep the HP-β-CD:SDS ratio of 1.75. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. FDR, false discovery rate; HP-β-CD, 2-hydroxypropyl-β-cyclodextrin; SCASP, SDS–cyclodextrin-assisted sample preparation; SDC, sodium deoxycholate; SWATH-MS, Sequential Window Acquisition of All Theoretical Mass Spectra.
Given HP-β-CD has much higher water solubility (>1200 mg/ml) than α-CD (145 mg/ml) and γ-CD (232 mg/ml) (28) and does not precipitate under acidic conditions, HP-β-CD was chosen in the subsequent experiments. We investigated several digestion factors in the SCASP method, such as different ratios of enzyme: substrate, diverse amount of starting material, varying incubation times, and trypsin or trypsin plus lysine-C digestions (Fig. 2, C–E and supplemental Table S1). These results demonstrated that SCASP exhibited similar performance with the SDC-based method. We also tested SCASP under different SDS concentrations. SCASP methods using 1%, 5%, and 10% SDS produced similar numbers of peptides and proteins (Fig. 2F and supplemental Table S1).
Comparison of SCASP With Other Sample Preparation Methods
Next, we compared SCASP to other SDS-depletion sample preparation methods, acetone precipitation, and FASP (9). Acetone precipitation is reported as a favored strategy among several SDS depletion techniques in a proteomics workflow (29). SCASP can identify 1655 proteins (the average number), which are 21% (360 of 1665) and 14% (233 of 1665) more than those identified by FASP and acetone precipitation, respectively (Fig. 3A and supplemental Table S1). About 7034 peptide ions (the average number) were identified using SCASP, which are the 56% (3948 of 7034) and 26% (1825 of 7034) increases compared with those identified by the FASP and acetone precipitation methods, respectively. CVs of proteins identified by FASP, acetone precipitation, and SCASP are 15.1, 6.2, and 2.3%, respectively, whereas the CVs of peptide ions are 21.1, 8.8, and 2.7%, respectively (supplemental Fig. S6A and supplemental Table S1). The CVs for peptides and proteins with the SDC method are 4.87 and 2.95%, respectively, which are comparable to those with SCASP. The CVs of protein intensities also showed similar pattern (supplemental Fig. S6B). The higher CVs of FASP and acetone precipitation methods than those of SCASP are expected, as both methods involved many more steps than SCASP. We also compared SCASP with SP3. About 5 μg proteins were processed by SCASP and SP3 methods in six biological replicates. SP3 was performed exactly following the protocol described by Hughes et al. (17). SCASP identified 5814 ± 309 precursors and 1402 ± 28 proteins, whereas SP3 identified 4757 ± 555 precursors and 1334 ± 107 proteins (supplemental Fig. S7 and supplemental Table S1). SCASP produced 18% more (1057 of 5814) precursors and 4.8% (68 of 1402) proteins than SP3 (supplemental Table S1).
Fig. 3.
Comparison of SCASP and other SDS-based methods.A, numbers of identified proteins and peptide ions from the starting material of 10 μg L929 lysate proteins using SCASP, acetone precipitation, and FASP. About 10 μg L929 lysate proteins were dissolved in 100 μl 1% SDS/100 mM Tris–HCl (pH 8.5), followed by SCASP, acetone precipitation, and FASP, respectively. After peptide cleanup, 1 μg peptide was analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. B, numbers of identified proteins and peptide ions from a whole-proteome analysis. About 100 μg HeLa proteins were processed by SCASP, the SDC method, and FASP, respectively. The peptides were fractionated using high-pH reverse phase LC. The fractions were analyzed by TripleTOF 5600. The MS data were searched by MSFragger, and identification results filtered at 1% protein FDR and 1% precursor FDR. C, overlap of peptide sequences and proteins produced by SCASP, the SDC method, and FASP. D, distribution of peptide length produced by SCASP, the SDC method, and FASP. E, GO analysis of identified proteins by SCASP, the SDC method, and FASP. FASP, filter-aided sample preparation; FDR, false discovery rate; GO, Gene Ontology; SCASP, SDS–cyclodextrin-assisted sample preparation; SDC, sodium deoxycholate.
To provide a systematic comparison of SCASP with other methods, we performed a whole-proteome scale analysis using SCASP, the SDC-based method, and FASP. About 100 μg proteins from HeLa cells were processed by these three methods. The peptides were subjected to high-pH reverse phase fractionation. The resulting 20 fractions were analyzed with DDA MS. In total, SCASP, iST, and FASP identified 110,707, 110,402, and 112,988 precursors, 8370, 8542, and 8430 proteins at 1% precursor, and protein FDR, respectively (Fig. 3B). About 38.8% of total peptide sequences (55,513 of 142,811) are shared by three methods, whereas 82.3% of total proteins (7639 of 9274) are commonly identified by three methods (Fig. 3C). Enabling match between run slightly enhanced overlapped peptides of three methods (supplemental Fig. S8). This result implied that trypsin exhibited different cleavage patterns under diverse conditions. We analyzed the hydrophobicity, charges, length, and missed cleavage of identified peptides. The peptides generated by three methods exhibited almost identical distribution of GRAVY values (supplemental Fig. S10A). The iST method produced more 2+ peptide ions than SCASP and FASP, whereas SCASP and FASP generated more 3+ and 4+ ones (supplemental Fig. S10B). Intriguing, SCASP and FASP, SDS-based methods, yielded longer length and more missed cleavages of peptides than the SDC-based method (Fig. 3D and supplemental Fig. S10C). Gene Ontology analysis showed that the numbers of the proteins that were clustered in the top 10 cellular localization by three methods are similar (Fig. 3E). These results demonstrated that there is no processing bias for SCASP method compared with the SDC or FASP method.
Application of SCASP in Processing Samples From Diverse Biological Sources
After processing the protein samples, we applied SCASP to other kinds of biological samples, such as cells or tissues. SCASP and SDC were used to digest seven human or murine cell lines, and the starting material is about 50,000 cells. For all tested cell lines, similar numbers of peptides and proteins were identified using SCASP and SDC methods (Fig. 4A and supplemental Table S1). Gene Ontology analysis showed that no significant cellular location difference was observed between the identified proteins by two methods (Fig. 4B). We further used SCASP, SDC, and urea methods to digest different numbers of 293T cells. SCASP and SDC methods produced nearly identical numbers of proteins, whereas the urea method produced significantly fewer proteins (Fig. 4C and supplemental Table S1). We noticed that SCASP produced 9% (975 of 10,860) more peptides than the SDC approach when taking 1 × 105 293T cells as the starting material. We compared the sequence coverage of common 1694 proteins identified by both methods and found SCASP generated more proteins with above 30% sequence coverage than the SDC-based approach (supplemental Fig. S11 and supplemental Table S1).
Fig. 4.
Comparison of SCASP and SDC-based methods on cell samples.A, numbers of identified proteins and peptide ions from 50,000 different cells using the SDC-based method and SCASP. About 50,000 cells were dissolved in 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers. The samples were boiled for 10 min. After peptide cleanup, the peptides corresponding to 1000 cells were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. B, GO analysis of proteins from seven cell lines using the SDC method or SCASP. Proteins were subjected to GO analysis (https://david.ncifcrf.gov/), and the percent of proteins that were assigned with “membrane” and “cytosol” was calculated. C, numbers of identified proteins and peptide ions from indicated different numbers of 293T using SCASP, the SDC-based method, and the urea method. Indicated numbers of 293T cells were dissolved in SDC, SDS, and urea buffers, respectively. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. D, numbers of identified proteins and peptide ions from indicated different numbers of HeLa cells using SCASP and the SDC-based method. Indicated numbers of 293T cells were dissolved in SDC and SDS buffers in max-recovery tubes, followed by digestion. About 1 μg peptides were analyzed by MS, and identified precursor ions and proteins were filtered at 1% FDR. CAA, 2-chloroacetamide; FDR, false discovery rate; GO, Gene Ontology; SCASP, SDS–cyclodextrin-assisted sample preparation; SDC, sodium deoxycholate; TCEP, Tris(2-carboxyethyl)phosphine hydrochloride.
To test the performance of SCAP to handle minimal samples, different numbers of HeLa cells were processed with SCASP. About 2 × 105 HeLa cells were first dissolved in 1% SDS, and the protein concentration was then determined by the bicinchoninic acid assay. About 45.6 μg of proteins can be extracted from 2 × 105 HeLa cells, which are close to 40 μg proteins assuming 200 pg per HeLa cell. Subsequently, 2 × 105 cells were diluted into 2 × 104, 1 × 104, and 5 × 103 cells, which are corresponding to 4.56, 2.28, and 1.14 μg proteins, respectively. These cells were processed by the SDC method and SCASP. About 1 μg peptides were analyzed by MS. For 45.6, 4.45, 2.28, and 1 μg starting material, nearly identical number of precursors were identified (Fig. 4D and supplemental Table S1). As for 4.45, 2.28, and 1 μg starting material, SCASP produced a slightly reduced number of proteins. The difference of identified proteins by both methods is within 10% (supplemental Table S1).
The yeast S. cerevisiae cells are protected by rigid cell walls, and it has been the general practice to use glass beads to impair yeast cell walls and help to release yeast cellular proteins (30). The plant cell wall comprises large amounts of cellulose and pectin and have a rigid secondary cell wall because of lignification of mature cells (31). It is generally accepted that plant samples were dissolved in SDS buffer followed by trichloroacetic acid/acetone precipitation. We attempted to use SCASP to process yeast S. cerevisiae cells and rice A. thaliana leaves without the use of the glass beads or trichloroacetic acid/acetone precipitation. The SCASP approach identified 1657 ± 6 proteins and 12,399 ± 141 peptides, whereas the SDC-based method yielded 1363 ± 30 proteins and 7270 ± 240 peptides (Fig. 5A and supplemental Table S1). For plant samples, 1777 ± 32 proteins and 7116 ± 190 peptides were identified using SCASP, whereas 1467 ± 56 proteins and 5660 ± 262 peptides were identified with the SDC-based method. Overall, the SCASP method produced 17% more proteins and 41% more peptides for yeast cell samples and 18% more proteins and 20% more peptides for rice leaf samples than the SDC-based method. We subsequently applied the SCASP method to seven murine tissue samples: heart, liver, small intestine, thymus, bladder, brain, and large intestine. SCASP can identify more proteins and peptides than SDC-based methods in seven tissues to varying degrees (Fig. 5B and supplemental Table S1). To further show the capability of SCASP to process the tissue samples, we used SCASP and SDC to digest FFPE murine tissues. SCASP identified about 10% more proteins and peptide ions than the SDC-based method, and the gain of peptides become more significant when the samples were analyzed on the more sensitive instrument (Fig. 5C and supplemental Table S1). We also used SCASP and the SDC-based methods to process the equal weight of human breast tumor tissues, and 1 μg peptides were analyzed by MS. SCASP identified 2154 ± 41 proteins and 10,493 ± 293 peptide ions, whereas the SDC-based method identified 2062 ± 24 and 9843 ± 321 peptide ions (Fig. 5D and supplemental Table S1). These results showed that SCASP can be applied in human tissue sample preparation for proteomics research.
Fig. 5.
Comparison of SCASP and SDC-based methods on yeast and tissue samples.A, numbers of identified proteins and peptide ions from yeast and rice leaf tissues using the SDC-based method and SCASP. About 107 yeast cells and 1 mg rice leaf powder were dissolved in 100 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers, respectively. The samples were boiled for 10 min. After peptide cleanup, around 2 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. B, numbers of identified proteins and peptide ions from seven different tissues using the SDC-based method and SCASP. About 10 mg of tissues were dissolved in 500 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers. After homogenization, 10 μg proteins were used for protein digestion. The samples were boiled for 10 min. After peptide cleanup, around 2 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. C, numbers of identified proteins and peptide ions from FFPE samples using the SDC-based method and SCASP. Deparaffinized FFPE samples were dissolved in 100 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers, respectively. The samples were boiled for 10 min. After peptide cleanup, around 2 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. D, numbers of identified proteins and peptide ions from human breast tumor tissues. About 10 mg of tissues were dissolved in 500 μl 1% SDS/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) or 1% SDC/10 mM TCEP/40 mM CAA/100 mM Tris–HCl (pH 8.5) buffers. After homogenization, 10 μg proteins were used for protein digestion. The samples were boiled for 10 min. After peptide cleanup, around 2 μg peptides were analyzed by MS. The identified peptide ions and proteins were filtered at 1% FDR. CAA, 2-chloroacetamide; FDR, false discovery rate; FFPE, formalin-fixed and paraffin-embedded; SCASP, SDS–cyclodextrin (CD)-assisted sample preparation; SDC, sodium deoxycholate; TCEP, Tris(2-carboxyethyl)phosphine hydrochloride.
Analysis of Dynamic Total and Membrane Proteome of VSV-Infected THP-1 Cells Using SCASP
Finally, we employed the SCASP method to investigate dynamic proteome and membrane proteome changes of VSV-infected human THP-1 cells. THP-1 cells were infected with VSV for five time points, and each experiment was conducted in biological duplicates (Fig. 6A). Membrane fractions and total proteins of THP-1 cells were digested and analyzed by ddaPASEF (parallel accumulation serial fragmentation) (32) and diaPASEF (33) MS. To build in-depth spectral library for targeted analysis of diaPASEF data, peptides from total proteins and membrane fractions were fractionated using high-pH LC. Single-run and fractionation ddaPASEF data were collectively searched with MSFragger (34), resulting in 9487 proteins and 396,778 peptide sequences at 1% protein and peptide FDR (supplemental Table S2). The precursor ions passing 1% protein FDR were used for generation of the spectral library by Skyline software (24). These spectral libraries were employed for targeted analysis of diaPASEF data. About 7031 proteins that contained 112,827 precursor ions were qualified across 20 diaPASEF runs at 1% FDR (q < 0.01), of which 6007 proteins were identified with at least two peptides (supplemental Table S2). Majority of human protein abundances of THP-1 cells did not change upon VSV stimulation (supplemental Fig. S12), and four VSV-encoding proteins were significantly upregulated (Fig. 6B). Correlation coefficient analysis of protein abundances between any two different samples revealed the excellent reproducibility of the entire experiments (supplemental Fig. S13). Principal component analysis showed that biological replicates were tightly clustered, and membrane samples and total protein samples were clearly separated (Fig. 6C). To identify VSV-regulated proteins, we performed differential expression analysis (supplemental Fig. S14). In total, 1230 proteins were downregulated, and 610 proteins were upregulated in THP-1 membrane fractions upon VSV stimulation. About 426 proteins were downregulated and 419 proteins were upregulated at THP-1 total protein levels upon VSV stimulation (supplemental Table S2). We manually checked extracted ion chromatography of all significantly changed peptides. Five proteins encoding by the VSV genome are upregulated markedly after VSV infection for 8 h (supplemental Fig. S15). Three human proteins were found to be induced under VSV infection (supplemental Fig. S16), of which RNA-binding protein FUS (FUS_HUMAN) was reported to be induced by interferon, which was stimulated by viral dsRNA (35). Four human proteins were downregulated by VSV stimulation (supplemental Fig. S16), of which PPP2R5D (2A5D_HUMAN) was degraded in T cells upon RNA virus HIV infection (36). The abundance of RAB18 increased in the membrane fraction, whereas it kept unchanged at the total protein level (Fig. 6D and supplemental Fig. S17). Considering RAB18 functions in mediating the formation of endoplasmic reticulum–lipid droplets (37), translocation of RAB18 from the cytosol into membrane revealed by our data suggested RAB18 may promote VSV virion assembly (38).
Fig. 6.
Application of SCASP in dynamic proteome profiling of VSV-infected THP-1 cells.A, experimental scheme of processing VSV-infected human monocyte THP cells for various time points using SCASP. B, heat map of quantified proteins in 20 THP-1 samples under VSV infection for the indicated time. Protein intensities were first normalized according to total protein intensities per run, and then log2 transformed. Fold change is represented by log2 values at indicated treated time minus those under control conditions. C, principal components analysis of protein intensities from 20 THP-1 samples. The biological replicates were labeled in the same color. D, product ions' XICs of peptide “LAIWDTAGQER” of human protein RAB18 in five THP-1 membrane samples and five lysate samples. CASP, SDS–cyclodextrin (CD)-assisted sample preparation; VSV, vesicular stomatitis virus; XIC, extracted ion chromatography.
Discussion
Development of simple and robust sample preparation methods is a hotspot question in proteomics research. SDS enables efficient protein extraction and protein denaturation, which makes SDS an ideal material for sample preparation. However, SDS removal is required before LC and MS analysis. Acetone precipitation is traditionally performed to remove SDS, which is time and labor extensive. FASP suffered sample loss when processing low microgram range of samples (39), whereas SDS precipitation with potassium chloride resulted in incomplete removal of SDS (29). Several protein-binding beads–based methods have recently been developed to deplete SDS in samples (10, 11, 12). These methods required additional commercial beads and involved multiple steps such as centrifugation and supernatant removal. In contrast, SCASP needs only one step prior to the addition of trypsin. In addition, the cost of SCASP is low if the local supplier of CD is available (see Experimental Procedures section). For protein samples, no significant difference was observed between SDC-based method and SCASP, which is probably attributed to the equal capability of SDC to dissolve membrane proteins relative to SDS (40, 41). However, the ability of SDS to disrupt intact cells or tissues is significantly better than SDC, especially for the biological materials, which are surrounded by rigid cell membranes. In this study, we observed that optimal performance of SCASP was achieved for yeast cells, plant tissues, and FFPE samples compared with the SDC-based method. It is anticipated that SCASP is highly suitable for preparation of lysis-resistant samples with limited starting amounts, such as metaproteomics (42) and FFPE samples–based clinical proteomics.
CDs have been employed for the removal of SDS in the peptide samples (16). It should be pointed out that our method differed clearly from that described in that study. In that study, CDs have not been removed in the peptide samples, which severely suppress the peptide signals. Furthermore, depletion of SDS at protein levels enabled the release of maximal trypsin activity compared with depletion of SDS at peptide levels.
CDs have been reported to interact with peptides or proteins (13); however, fundamental studies on the interactions of CDs with proteins have been scarce presumably because of very weak interaction of CDs with proteins (43). For example, the interaction of CDs with human and bovine serum albumins, respectively, was investigated (44), and steady state fluorescence, circular dichroism spectra, and isothermal titration calorimetric data showed no significant interaction. Indeed, CDs interact with the hydrophobic amino acids on the surface of the protein, and the interaction of CDs with these residues may be perturbed significantly by the environments (43). We did not observe a significant difference between SCASP and the SDC-based method in performing digestion of minimal samples or the in-depth whole-proteome analysis. Although we cannot conclude that no peptides or proteins are removed along with CDs, the limited peptides or proteins go away with CDs during the SCASP method.
In summary, we presented an SDS-based sample preparation method for proteomics research, which is simple, robust, and reproducible. SCASP can be applied for various kinds of samples, especially for challenging samples, such as yeast cells, animal, or plant tissues.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (45) with the data set identifier PXD023557.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Science Foundation of China for Fostering Talents in Basic Research (J1310027) and the Fundamental Research Funds for the Central Universities (20720190087). We thank Dr Chuanchuang Xie and Ms Yaying Wu for helping with MS data acquisition.
Author contributions
G. G. performed the experiments under C.- Q. Z. instruction; X. X, X.- F. Z., and J. W. provided clinical human breast cancer samples; X. C. provided plant leaf and FFPE samples; C.- Q. Z. conceived the concept, performed MS analysis, analyzed MS data, and wrote the article.
Supplemental Data
References
- 1.Kachuk C., Doucette A.A. The benefits (and misfortunes) of SDS in top-down proteomics. J. Proteomics. 2018;175:75–86. doi: 10.1016/j.jprot.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Rundlett K.L., Armstrong D.W. Mechanism of signal suppression by anionic surfactants in capillary electrophoresis-electrospray ionization mass spectrometry. Anal. Chem. 1996;68:3493–3497. doi: 10.1021/ac960472p. [DOI] [PubMed] [Google Scholar]
- 3.Botelho D., Wall M.J., Vieira D.B., Fitzsimmons S., Liu F., Doucette A. Top-down and bottom-up proteomics of SDS-containing solutions following mass-based separation. J. Proteome Res. 2010;9:2863–2870. doi: 10.1021/pr900949p. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld J., Capdevielle J., Guillemot J.C., Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 5.Sun D., Wang N., Li L. Integrated SDS removal and peptide separation by strong-cation exchange liquid chromatography for SDS-assisted shotgun proteome analysis. J. Proteome Res. 2012;11:818–828. doi: 10.1021/pr200676v. [DOI] [PubMed] [Google Scholar]
- 6.Mechin V., Damerval C., Zivy M. Total protein extraction with TCA-acetone. Methods Mol. Biol. 2007;355:1–8. doi: 10.1385/1-59745-227-0:1. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J.Y., Dann G.P., Shi T., Wang L., Gao X., Su D., Nicora C.D., Shukla A.K., Moore R.J., Liu T., Camp D.G., 2nd, Smith R.D., Qian W.J. Simple sodium dodecyl sulfate-assisted sample preparation method for LC-MS-based proteomics applications. Anal. Chem. 2012;84:2862–2867. doi: 10.1021/ac203394r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manza L.L., Stamer S.L., Ham A.J., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 9.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 10.Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zougman A., Selby P.J., Banks R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics. 2014;14:1006–1010. doi: 10.1002/pmic.201300553. [DOI] [PubMed] [Google Scholar]
- 12.Batth T.S., Tollenaere M.A.X., Ruther P., Gonzalez-Franquesa A., Prabhakar B.S., Bekker-Jensen S., Deshmukh A.S., Olsen J.V. Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol. Cell. Proteomics. 2019;18:1027–1035. doi: 10.1074/mcp.TIR118.001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uekama K., Hirayama F., Irie T. Cyclodextrin drug carrier systems. Chem. Rev. 1998;98:2045–2076. doi: 10.1021/cr970025p. [DOI] [PubMed] [Google Scholar]
- 14.Davis M.E., Brewster M.E. Cyclodextrin-based pharmaceutics: Past, present and future. Nat. Rev. Drug Discov. 2004;3:1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 15.Turco Liveri V., C G., Giammona G., Pitarresi G., Puglisi G., Ventura C. Calorimetric investigation of the complex formation between surfactants and α-, β- and γ-cyclodextrins. Thermochim. Acta. 1992;199:125–132. [Google Scholar]
- 16.Quirino J.P. Sodium dodecyl sulfate removal during electrospray ionization using cyclodextrins as simple sample solution additive for improved mass spectrometric detection of peptides. Anal. Chim. Acta. 2018;1005:54–60. doi: 10.1016/j.aca.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Hughes C.S., Moggridge S., Muller T., Sorensen P.H., Morin G.B., Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 18.Lai X. Reproducible method to enrich membrane proteins with high purity and high yield for an LC-MS/MS approach in quantitative membrane proteomics. Electrophoresis. 2013;34:809–817. doi: 10.1002/elps.201200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Veiga Leprevost F., Haynes S.E., Avtonomov D.M., Chang H.Y., Shanmugam A.K., Mellacheruvu D., Kong A.T., Nesvizhskii A.I. Philosopher: A versatile toolkit for shotgun proteomics data analysis. Nat. Methods. 2020;17:869–870. doi: 10.1038/s41592-020-0912-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D., Gan G., Chen X., Zhong C.Q. QuantPipe: A user-friendly pipeline software tool for DIA data analysis based on the OpenSWATH-PyProphet-TRIC workflow. J. Proteome Res. 2021;20:1096–1102. doi: 10.1021/acs.jproteome.0c00704. [DOI] [PubMed] [Google Scholar]
- 21.Rost H.L., Rosenberger G., Navarro P., Gillet L., Miladinovic S.M., Schubert O.T., Wolski W., Collins B.C., Malmstrom J., Malmstrom L., Aebersold R. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberger G., Bludau I., Schmitt U., Heusel M., Hunter C.L., Liu Y., MacCoss M.J., MacLean B.X., Nesvizhskii A.I., Pedrioli P.G.A., Reiter L., Rost H.L., Tate S., Ting Y.S., Collins B.C. Statistical control of peptide and protein error rates in large-scale targeted data-independent acquisition analyses. Nat. Methods. 2017;14:921–927. doi: 10.1038/nmeth.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rost H.L., Liu Y., D'Agostino G., Zanella M., Navarro P., Rosenberger G., Collins B.C., Gillet L., Testa G., Malmstrom L., Aebersold R. TRIC: An automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods. 2016;13:777–783. doi: 10.1038/nmeth.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polpitiya A.D., Qian W.J., Jaitly N., Petyuk V.A., Adkins J.N., Camp D.G., 2nd, Anderson G.A., Smith R.D. DAnTE: A statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 27.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 28.Gidwani B., Vyas A. A comprehensive review on cyclodextrin-based carriers for delivery of chemotherapeutic cytotoxic anticancer drugs. Biomed. Res. Int. 2015;2015:198268. doi: 10.1155/2015/198268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kachuk C., Stephen K., Doucette A. Comparison of sodium dodecyl sulfate depletion techniques for proteome analysis by mass spectrometry. J. Chromatogr. A. 2015;1418:158–166. doi: 10.1016/j.chroma.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 30.von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS One. 2007;2 doi: 10.1371/journal.pone.0001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W., Scali M., Vignani R., Spadafora A., Sensi E., Mazzuca S., Cresti M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003;24:2369–2375. doi: 10.1002/elps.200305500. [DOI] [PubMed] [Google Scholar]
- 32.Meier F., Brunner A.D., Koch S., Koch H., Lubeck M., Krause M., Goedecke N., Decker J., Kosinski T., Park M.A., Bache N., Hoerning O., Cox J., Rather O., Mann M. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics. 2018;17:2534–2545. doi: 10.1074/mcp.TIR118.000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier F., Brunner A.-D., Frank M., Ha A., Bludau I., Voytik E., Kaspar-Schoenefeld S., Lubeck M., Raether O., Aebersold R., Collins B.C., Röst H.L., Mann M. Parallel accumulation – serial fragmentation combined with data-independent acquisition (diaPASEF): Bottom-up proteomics with near optimal ion usage. bioRxiv. 2020 doi: 10.1101/656207. [DOI] [PubMed] [Google Scholar]
- 34.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelkovnikova T.A., An H., Skelt L., Tregoning J.S., Humphreys I.R., Buchman V.L. Antiviral immune response as a trigger of FUS proteinopathy in amyotrophic lateral sclerosis. Cell Rep. 2019;29:4496–4508.e4494. doi: 10.1016/j.celrep.2019.11.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenwood E.J., Matheson N.J., Wals K., van den Boomen D.J., Antrobus R., Williamson J.C., Lehner P.J. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. Elife. 2016;5 doi: 10.7554/eLife.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D., Zhao Y.G., Li D., Zhao H., Huang J., Miao G., Feng D., Liu P., Li D., Zhang H. The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep. 2019;27:343–358.e345. doi: 10.1016/j.celrep.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 38.Salloum S., Wang H., Ferguson C., Parton R.G., Tai A.W. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sielaff M., Kuharev J., Bohn T., Hahlbrock J., Bopp T., Tenzer S., Distler U. Evaluation of FASP, SP3, and iST protocols for proteomic sample preparation in the low microgram range. J. Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 40.Lin Y., Zhou J., Bi D., Chen P., Wang X., Liang S. Sodium-deoxycholate-assisted tryptic digestion and identification of proteolytically resistant proteins. Anal. Biochem. 2008;377:259–266. doi: 10.1016/j.ab.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J., Zhou T., Cao R., Liu Z., Shen J., Chen P., Wang X., Liang S. Evaluation of the application of sodium deoxycholate to proteomic analysis of rat hippocampal plasma membrane. J. Proteome Res. 2006;5:2547–2553. doi: 10.1021/pr060112a. [DOI] [PubMed] [Google Scholar]
- 42.Wilmes P., Heintz-Buschart A., Bond P.L. A decade of metaproteomics: Where we stand and what the future holds. Proteomics. 2015;15:3409–3417. doi: 10.1002/pmic.201500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurarslan A., Joijode A., Shen J., Narayanan G., Antony G.J., Li S., Caydamli Y., Tonelli A.E. Reorganizing polymer chains with cyclodextrins. Polymers (Basel) 2017;9:673. doi: 10.3390/polym9120673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oi W., Isobe M., Hashidzume A., Harada A. Macromolecular recognition: Discrimination between human and bovine serum albumins by cyclodextrins. Macromol. Rapid Commun. 2011;32:501–505. doi: 10.1002/marc.201000699. [DOI] [PubMed] [Google Scholar]
- 45.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., Hermjakob H., Zhu Y. iProX: An integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (45) with the data set identifier PXD023557.