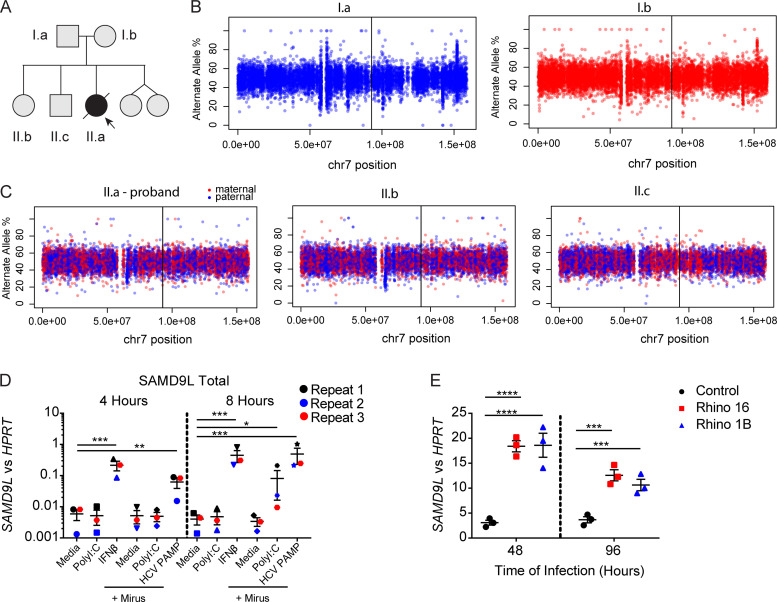
Figure S1.
Chromosomal analysis of the proband and unaffected relatives and regulation of SAMD9L expression. (A) Pedigree from the family annotated. (B and C) Allele frequency of phased heterozygous variants on chromosome (chr) 7 as calculated from the allelic read depth from WGS data. (C) The variants are color coded based on the parental origin for siblings from the respective parents shown in B. The vertical lines indicate the position of the SAMD9L proband variant (GenBank accession no. NM_152703.2:c.2658_2659del). (D) A549 lung epithelial cells were treated with media alone (1.25 µg/ml), high-molecular-weight poly(I:C), or human IFN-β (100 U/ml) or transfected with 1.25 µg high-molecular-weight poly(I:C) or 1 μg hepatitis C virus pathogen-associated molecular pattern using Mirus Bio lipofection reagents following the manufacturer protocol for 4 or 8 h before isolation of mRNA. RT-PCR for SAMD9L mRNA transcripts compared with HPRT mRNA transcript means ± SD from n = 3 pooled experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA with Tukey’s multiple comparison test). (E) Ex vivo–cultured lower airway epithelial cells from healthy children (n = 3) were cultured at baseline and during infection with HRV-A16 or -1B (Rhino 16 and 1B, respectively) for 48 and 96 h before mRNA isolation. Representative from n = 2 independent experiments (***, P < 0.001; ****, P < 0.0001 by one-way ANOVA and Tukey’s multiple comparison test).