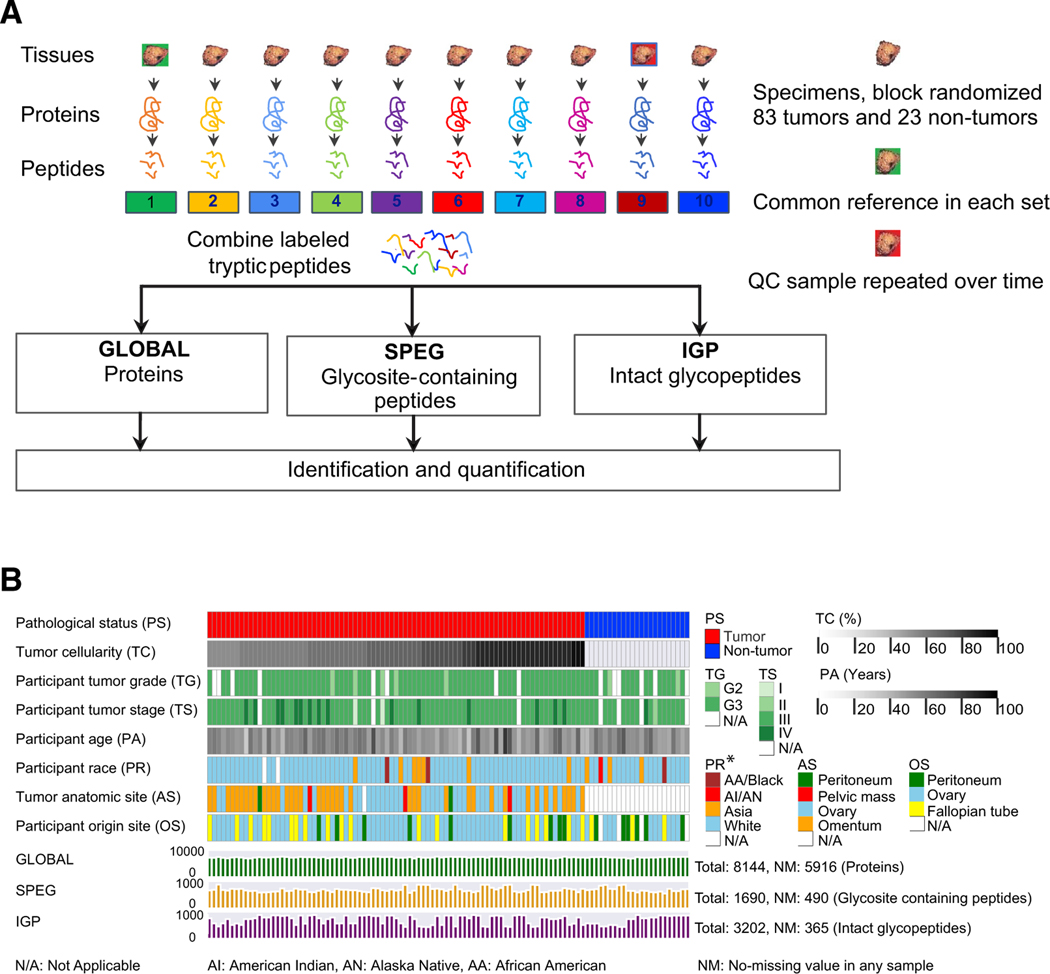
Figure 1. The Workflow of the Integrated Glycoproteomic Strategy to Analyze HGSCs and Non-tumor Tissues.
(A) Proteins from 83 HGSC tumor tissues, 23 non-tumor tissues, pooled reference sample, and technical replicates of quality control sample were digested by trypsin to peptides, which were labeled by TMT and analyzed by global proteomic analysis (GLOBAL), as well as glycosite-containing peptides (SPEG), and intact glycopeptides (IGPs) analysis using LC-MS/MS.
(B) The clinical phenotypes and data profiling of proteomic (GLOBAL) and glycoproteomic (SPEG and IGP) data of 83 tumor and 23 non-tumor tissues.