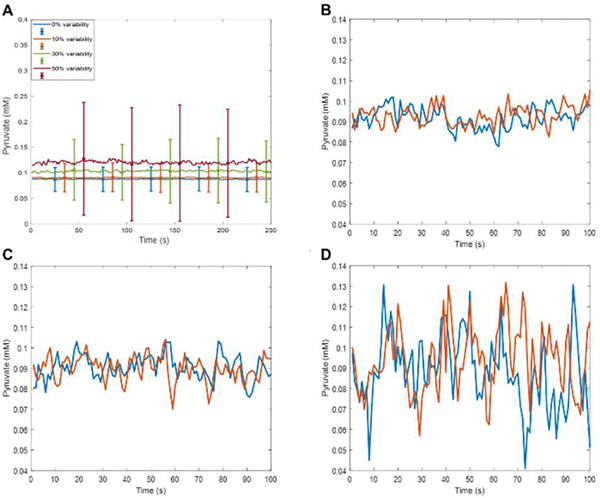
Fig. 3.
Effect of random variability of initial concentrations of all glycolytic intermediates and kinetic constants on the simulated concentration of pyruvate. A) Means and standard deviations calculated over populations of 100 independent cells with variability applied to the concentrations of the metabolites of interest and the enzymes involved in all of the glycolysis reactions. The percent variability is derived from a zero mean Gaussian with a standard deviation of x/3 and is multiplied by the nominal concentration value. The initial concentrations of the metabolites of interest are chosen that way, and for the concentration of other enzymes involved, the operation is repeated every second, while the simulation step is 0.1 millisecond. Computation time for simulating 100 cells for 300 seconds with 0.1 millisecond time step was 36,767.18 seconds. B) Two individual cells, 10% variability of initial concentrations and 10% Gaussian noise. C) Two individual cells, 20% variability of initial concentrations and 20% Gaussian noise. D) Two individual cells, 50% variability of initial concentrations and 50% Gaussian noise.