Figure 2. The ubiquitin-proteasome pathway influences proteostasis, the cell cycle, and filamentation.
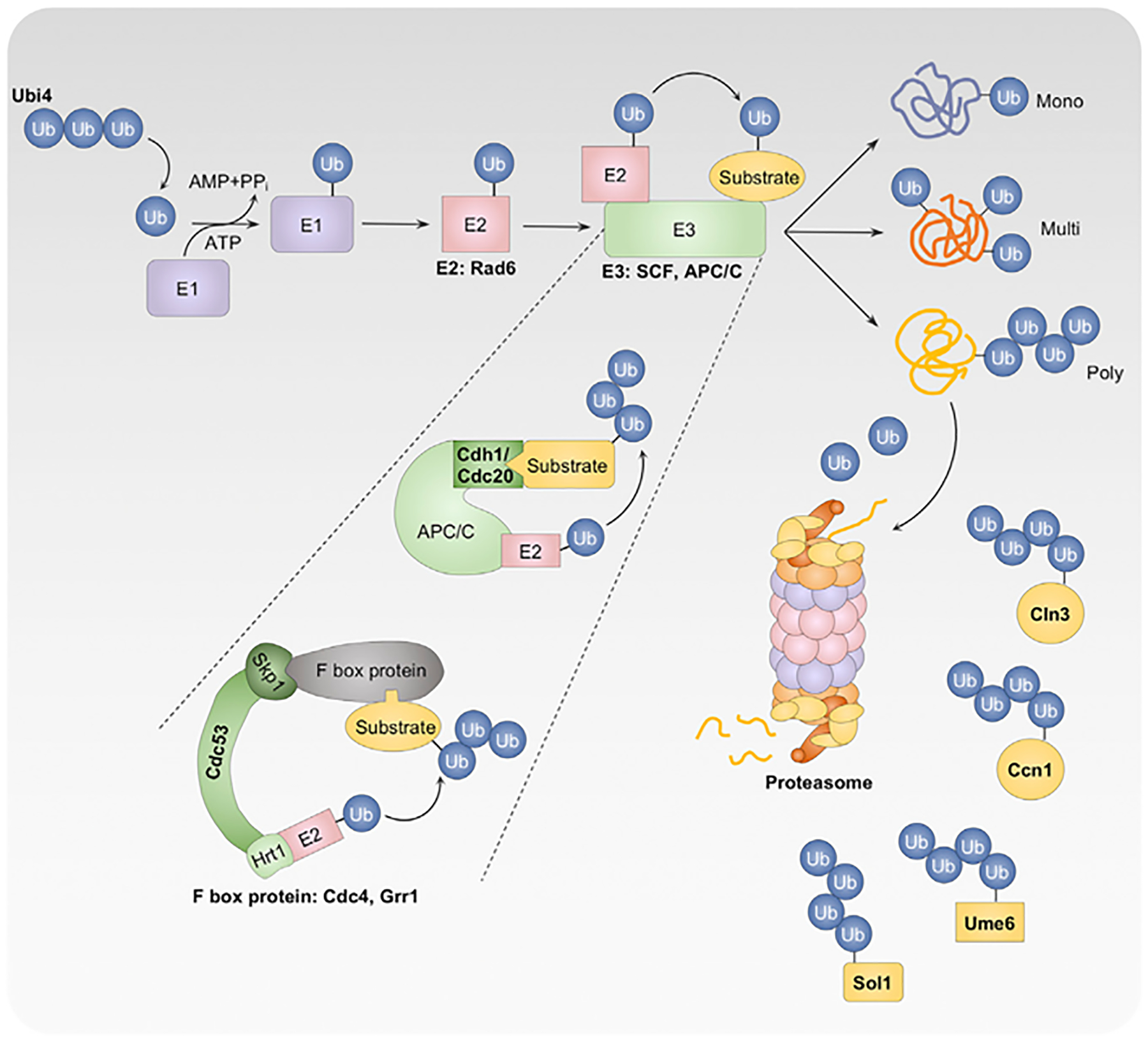
The polyubiquitin Ubi4 is cleaved to generate ubiquitin monomers. Ubiquitin is activated by a ubiquitin-activating enzyme (E1) in an ATP-dependent manner; transferred to a ubiquitin-conjugating enzyme (E2), such as the putative E2 Rad6; and subsequently covalently attached to substrate proteins by a ubiquitin ligase (E3). E3 is responsible for substrate selection and specificity. Addition of a single ubiquitin molecule to a substrate generates monoubiquitinated proteins, whereas multiple rounds of ubiquitination generate multi- or polyubiquitinated proteins. Polyubiquitinated proteins are preferentially targeted to the proteasome for degradation. The Skp1-Cullin-F-box (SCF) complex (consisting of the linker protein Skp1, scaffold protein Cdc53, and F-box protein) and the anaphase-promoting complex/cyclosome (APC/C) complex (including the coactivators Cdc20 and Cdh1) are important for proteasome-mediated turnover of proteins involved in the cell cycle and filamentation. UPP proteins and substrates with known roles in regulating C. albicans filamentation are indicated in bold.
