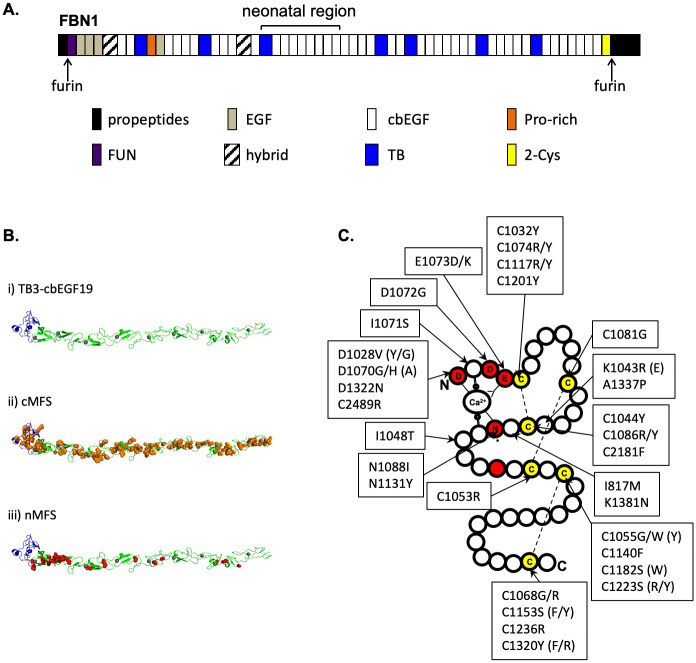
Fig 1. FBN1 domain organisation and sites of neonatal Marfan syndrome mutations.
A) The structure of FBN1 is dominated by cbEGF domains interspersed with TB domains. The majority of nMFS-associated mutations affect the central TB3- cbEGF18 domains, which have traditionally been referred to as the "neonatal region". B) A model of the neonatal region (i) was created using Modeller software [35] and coordinates from the structures of domains TB4 [4], cbEGF12-13 [36] and cbEGF32-33 [37]. TB3 is coloured blue, cbEGFs are coloured green with Ca2+ ions shown as grey spheres. Substitutions associated with cMFS (panel ii, orange) are found evenly distributed across the neonatal region while those associated with nMFS (panel iii, red) show a clustering around cbEGF11-12. C) Plotting the positions of known nMFS-associated substitutions on a schematic representation of a cbEGF domain shows that in many cases the effect of the substitution is structural, affecting either cysteines involved in disulphide bond formation (yellow) or Ca2+-binding consensus residues (red). Where nMFS and cMFS substitutions have been found at the same position, the cMFS version is shown in brackets. Data extracted from the Universal Mutation Database (http://umd.be/FBN1) [38].