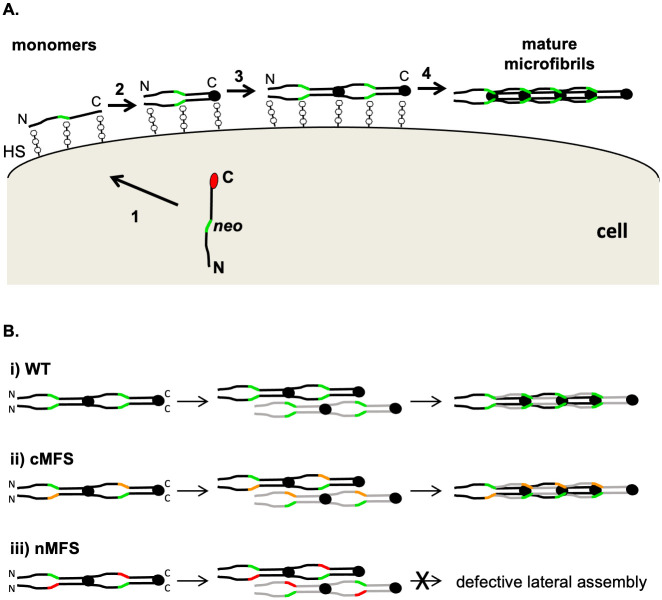
Fig 6. Model of the FBN1 neonatal region’s roles in microfibril assembly and nMFS pathogenesis.
A) In normal FBN1 assembly, secretion of the protein is coupled to the proteolytic cleavage (1) of the C-terminal propeptide (red circle). FBN1 interactions with heparan sulphate proteoglycans (HS), at sites including the N- and C-termini and neonatal region (other sites not shown), limit diffusion from the cell surface during assembly. Multimerisation of C-terminal domains, previously blocked by the propeptide, initiate the assembly process (2) and promote intermolecular avidity-driven interactions with N-terminal domains (3) that result in a head-to-tail alignment of monomers. The neonatal region (green) may be involved in the higher order, lateral assembly of FBN1 to form the mature microfibril (4). B) Lateral assembly (panel A, step 4) would occur between FBN1 molecules (black and grey) that have already undergone C-terminal multimerisation and head-to-tail alignment. Compared to the wild type case (i), cMFS-associated mutations in the neonatal region (ii; orange) appear to incorporate into microfibrils at near-normal levels. In contrast, mutations affecting the neonatal region that lead to nMFS (iii; red) are unlikely to affect N- to C-terminal interactions but would disrupt later stages of assembly such as lateral association. Mutants such as ΔcbEGF16 may not directly affect the neonatal assembly site, but would affect the position and register of the region in the maturing microfibril. In addition to preventing the incorporation of mutant molecules into microfibrils, the interaction between wild type and mutant variants at step 3 would reduce the amount of wild type FBN1 deposited into the matrix, resulting in microfibril levels that are less than expected for haploinsufficiency.